Delivery determinants of an Acinetobacter baumannii type VI secretion system bifunctional peptidoglycan hydrolase
- PMID: 39745415
- PMCID: PMC11796386
- DOI: 10.1128/mbio.02627-24
Delivery determinants of an Acinetobacter baumannii type VI secretion system bifunctional peptidoglycan hydrolase
Abstract
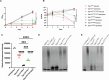
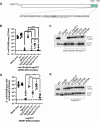
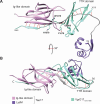
Acinetobacter baumannii is a Gram-negative opportunistic pathogen and is a common cause of nosocomial infections. The increasing development of antibiotic resistance in this organism is a global health concern. The A. baumannii clinical isolate AB307-0294 produces a type VI secretion system (T6SS) that delivers three antibacterial effector proteins that give this strain a competitive advantage against other bacteria in polymicrobial environments. Each effector, Tse15, Tde16, and Tae17, is delivered via a non-covalent interaction with a specific T6SS VgrG protein (VgrG15, VgrG16, and VgrG17, respectively). Here we define the regions of interaction between Tae17 and its cognate delivery protein VgrG17 and identify that amino acids G1069 and W1075 in VgrG17 are essential for Tae17 delivery via the T6SS, the first time such specific delivery determinants of T6SS cargo effectors have been defined. Furthermore, we determine that the Tae17 effector is a multidomain, bifunctional, peptidoglycan-degrading enzyme that has both amidase activity, which targets the sugar-peptide bonds, and lytic transglycosylase activity, which targets the peptidoglycan sugar backbone. Moreover, we show that the Tae17 transglycosylase activity is more important than amidase activity for the killing of Escherichia coli. This study provides molecular insight into how the T6SS allows A. baumannii strains to gain dominance in polymicrobial communities and thus improve their chances of survival and transmission.IMPORTANCEWe have shown that the Acinetobacter baumannii T6SS effector Tae17 is a modular, bifunctional, peptidoglycan-degrading enzyme that has both lytic transglycosylase and amidase activities. Both activities contribute to the ability to degrade peptidoglycan, but the transglycosylase activity was more important for the killing of Escherichia coli. We have defined the specific regions of Tae17 and its cognate delivery protein VgrG17 that are necessary for the non-covalent interactions and, for the first time, identified specific amino acids essential for T6SS cargo effector delivery. This work contributes to our molecular understanding of bacterial competition strategies in polymicrobial environments and may provide a window to design new therapeutic approaches for combating infection by A. baumannii.
Keywords: Acinetobacter baumannii; amidase; lytic transglycosylase; peptidoglycan hydrolase; toxic effector; type VI secretion system.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, Pulcini C, Kahlmeter G, Kluytmans J, Carmeli Y, Ouellette M, Outterson K, Patel J, Cavaleri M, Cox EM, Houchens CR, Grayson ML, Hansen P, Singh N, Theuretzbacher U, Magrini N, WHO Pathogens Priority List Working Group . 2018. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 18:318–327. doi:10.1016/S1473-3099(17)30753-3 - DOI - PubMed
-
- Russell AB, Singh P, Brittnacher M, Bui NK, Hood RD, Carl MA, Agnello DM, Schwarz S, Goodlett DR, Vollmer W, Mougous JD. 2012. A widespread bacterial type VI secretion effector superfamily identified using A heuristic approach. Cell Host Microbe 11:538–549. doi:10.1016/j.chom.2012.04.007 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

