A Clostridioides difficile cell-free gene expression system for prototyping and gene expression analysis
- PMID: 39745467
- PMCID: PMC11784378
- DOI: 10.1128/aem.01566-24
A Clostridioides difficile cell-free gene expression system for prototyping and gene expression analysis
Abstract
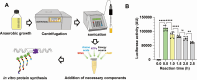
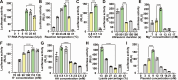
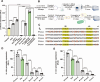
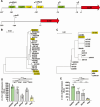
Clostridioides difficile is an obligate anaerobic, Gram-positive bacterium that produces toxins. Despite technological progress, conducting gene expression analysis of C. difficile under different conditions continues to be labor-intensive. Therefore, there is a demand for simplified tools to investigate the transcriptional and translational regulation of C. difficile. The cell-free gene expression (CFE) system has demonstrated utility in various applications, including prototyping, protein production, and in vitro screening. In this study, we developed a C. difficile CFE system capable of in vitro transcription and translation (TX-TL) in the presence of oxygen. Through optimization of cell extract preparation and reaction systems, we increased the protein yield significantly. Furthermore, our observations indicated that this system exhibited higher protein yield using linear DNA templates than circular plasmids for in vitro expression. The prototyping capability of the C. difficile CFE system was assessed using a series of synthetic Clostridium promoters, demonstrating a good correlation between in vivo and in vitro expression. Additionally, we tested the expression of tcdB and tcdR from clinically relevant C. difficile strains using the CFE system, confirming higher toxin expression of the hypervirulent strain R20291. We believe that the CFE system can not only serve as a platform for in vitro protein synthesis and genetic part prototyping but also has the potential to be a simplified model for studying metabolic regulations in Clostridioides difficile.IMPORTANCEClostridioides difficile has been listed as an urgent threat due to its antibiotic resistance, and it is crucial to conduct gene expression analysis to understand gene functionality. However, this task can be challenging, given the need to maintain the bacterium in an anaerobic environment and the inefficiency of introducing genetic material into C. difficile cells. Conversely, the C. difficile cell-free gene expression (CFE) system enables in vitro transcription and translation in the presence of oxygen within just half an hour. Furthermore, the composition of the CFE system is adaptable, permitting the addition or removal of elements, regulatory proteins for example, during the reaction. As a result, this system could potentially offer an efficient and accessible approach to accelerate the study of gene expression and function in Clostridioides difficile.
Keywords: CFE system; Clostridioides difficile; in vitro expression; prototyping; tcdR and tcdB expression.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

