Transcriptomic Profiles in Nasal Epithelium and Asthma Endotypes in Youth
- PMID: 39745770
- PMCID: PMC11775736
- DOI: 10.1001/jama.2024.22684
Transcriptomic Profiles in Nasal Epithelium and Asthma Endotypes in Youth
Abstract
Importance: T helper 2 (T2) cells and T helper 17 (T17) cells are CD4+ T cell subtypes involved in asthma. Characterizing asthma endotypes based on these cell types in diverse groups is important for developing effective therapies for youths with asthma.
Objective: To identify asthma endotypes in school-aged youths aged 6 to 20 years by examining the distribution and characteristics of transcriptomic profiles in nasal epithelium.
Design, setting, and participants: Cross-sectional analysis of nasal epithelial samples from 3 studies of youths with asthma aged 6 to 20 years: Stress and Treatment Response in Puerto Rican and African American Children with Asthma (STAR; N = 156), Epigenetic Variation and Childhood Asthma in Puerto Ricans (EVA-PR; N = 237), and Vitamin D Kids Asthma (VDKA; N = 66).
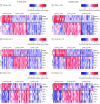
Main outcomes and measures: The primary outcome was nasal epithelial transcription profiles of 3 T2 and 5 T17 pathway genes. Clinical characteristics, total and allergen-specific immunoglobulin E (IgE), blood eosinophils, and lung function were compared across profiles in all studies.
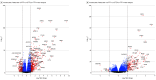
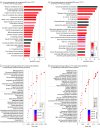
Results: Mean ages for STAR, EVA-PR, and VDKA participants were 14.2, 15.4, and 10.3 years, respectively. The percentage of female participants ranged from 41% to 53.2% across studies. The predominant race or ethnicity was Puerto Rican in EVA-PR (100%) and Black or African American in STAR (71.8%) and VDKA (57.6%). Three transcriptomic profiles were identified: high T2 expression (T2HIGH), high T17 expression (T17HIGH), and low expression of both pathways (T2LOW/T17LOW). Across studies, T2HIGH was present in 23% to 29% of participants, T17HIGH in 35% to 47%, and T2LOW/T17LOW in 30% to 38%. In each study, median total IgE and blood eosinophils for the T2HIGH profile was higher than for the T2LOW profiles (IgE, 584-869 vs 105-382 IU/mL; eosinophils, 343-560 vs 164-413 cells/mL). Of the participants in all profiles, at least 50% had 1 or more positive allergen-specific IgEs. A differential expression meta-analysis identified 3516 and 2494 differentially expressed genes for the T2HIGH and T17HIGH profiles, respectively. The T17HIGH profile was associated with interleukin 17 and neutrophil signaling pathways and the T2HIGH profile was associated with interleukin 13 signaling pathways.
Conclusions and relevance: Nasal transcriptomic profiles consistent with T2-high, T17-high, and T2-low/T17-low endotypes occurred in similar proportions across 3 studies of predominantly racially and ethnically minoritized youths with asthma. Most participants had T2-low asthma endotypes and sensitization to 1 or more allergens was common among these endotypes.
Conflict of interest statement
Figures

Comment on
-
Uncovering Transcriptional Endotypes in Pediatric Asthma-A New Lens on Biomarker-Based Clinical Stratification.JAMA. 2025 Jan 28;333(4):293-294. doi: 10.1001/jama.2024.26977. JAMA. 2025. PMID: 39745764 No abstract available.
References
-
- US Centers for Disease Control and Prevention . Data, statistics, and surveillance—asthma surveillance data. Reviewed July 24, 2024. Accessed June 25, 2024. https://www.cdc.gov/asthma/asthmadata.htm
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

