ATG9A facilitates the closure of mammalian autophagosomes
- PMID: 39745851
- PMCID: PMC11694768
- DOI: 10.1083/jcb.202404047
ATG9A facilitates the closure of mammalian autophagosomes
Abstract
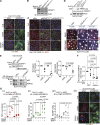
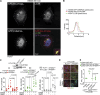
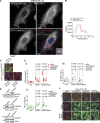
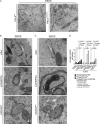
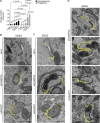
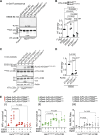
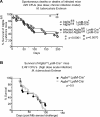
Canonical autophagy captures within specialized double-membrane organelles, termed autophagosomes, an array of cytoplasmic components destined for lysosomal degradation. An autophagosome is completed when the growing phagophore undergoes ESCRT-dependent membrane closure, a prerequisite for its subsequent fusion with endolysosomal organelles and degradation of the sequestered cargo. ATG9A, a key integral membrane protein of the autophagy pathway, is best known for its role in the formation and expansion of phagophores. Here, we report a hitherto unappreciated function of mammalian ATG9A in directing autophagosome closure. ATG9A partners with IQGAP1 and key ESCRT-III component CHMP2A to facilitate this final stage in autophagosome formation. Thus, ATG9A is a central hub governing all major aspects of autophagosome membrane biogenesis, from phagophore formation to its closure, and is a unique ATG factor with progressive functionalities affecting the physiological outputs of autophagy.
© 2025 Javed et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures






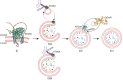
Similar articles
-
ATG9A controls all stages of autophagosome biogenesis.Autophagy. 2025 Aug;21(8):1859-1861. doi: 10.1080/15548627.2025.2494802. Epub 2025 Apr 27. Autophagy. 2025. PMID: 40241347
-
Blocking autophagosome closure manifests the roles of mammalian Atg8-family proteins in phagophore formation and expansion during nutrient starvation.Autophagy. 2025 May;21(5):1059-1074. doi: 10.1080/15548627.2024.2443300. Epub 2025 Jan 20. Autophagy. 2025. PMID: 39694607 Free PMC article.
-
Temporal dissection of the roles of Atg4 and ESCRT in autophagosome formation in yeast.Cell Death Differ. 2025 May;32(5):866-879. doi: 10.1038/s41418-024-01438-8. Epub 2024 Dec 23. Cell Death Differ. 2025. PMID: 39715823
-
Emerging roles of ATG9/ATG9A in autophagy: implications for cell and neurobiology.Autophagy. 2024 Nov;20(11):2373-2387. doi: 10.1080/15548627.2024.2384349. Epub 2024 Aug 4. Autophagy. 2024. PMID: 39099167 Free PMC article. Review.
-
ATG9 Not Just an Autophagy Related Protein.J Mol Biol. 2025 Sep 15;437(18):169288. doi: 10.1016/j.jmb.2025.169288. Epub 2025 Jun 11. J Mol Biol. 2025. PMID: 40513646 Review.
References
-
- Axe, E.L., Walker S.A., Manifava M., Chandra P., Roderick H.L., Habermann A., Griffiths G., and Ktistakis N.T.. 2008. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 182:685–701. 10.1083/jcb.200803137 - DOI - PMC - PubMed
-
- Bertin, A., de Franceschi N., de la Mora E., Maity S., Alqabandi M., Miguet N., di Cicco A., Roos W.H., Mangenot S., Weissenhorn W., and Bassereau P.. 2020. Human ESCRT-III polymers assemble on positively curved membranes and induce helical membrane tube formation. Nat. Commun. 11:2663. 10.1038/s41467-020-16368-5 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

