The Transformative Potential of Large Language Models in Mining Electronic Health Records Data: Content Analysis
- PMID: 39746191
- PMCID: PMC11739723
- DOI: 10.2196/58457
The Transformative Potential of Large Language Models in Mining Electronic Health Records Data: Content Analysis
Abstract
Background: In this study, we evaluate the accuracy, efficiency, and cost-effectiveness of large language models in extracting and structuring information from free-text clinical reports, particularly in identifying and classifying patient comorbidities within oncology electronic health records. We specifically compare the performance of gpt-3.5-turbo-1106 and gpt-4-1106-preview models against that of specialized human evaluators.
Objective: We specifically compare the performance of gpt-3.5-turbo-1106 and gpt-4-1106-preview models against that of specialized human evaluators.
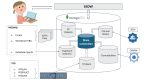
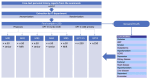
Methods: We implemented a script using the OpenAI application programming interface to extract structured information in JavaScript object notation format from comorbidities reported in 250 personal history reports. These reports were manually reviewed in batches of 50 by 5 specialists in radiation oncology. We compared the results using metrics such as sensitivity, specificity, precision, accuracy, F-value, κ index, and the McNemar test, in addition to examining the common causes of errors in both humans and generative pretrained transformer (GPT) models.
Results: The GPT-3.5 model exhibited slightly lower performance compared to physicians across all metrics, though the differences were not statistically significant (McNemar test, P=.79). GPT-4 demonstrated clear superiority in several key metrics (McNemar test, P<.001). Notably, it achieved a sensitivity of 96.8%, compared to 88.2% for GPT-3.5 and 88.8% for physicians. However, physicians marginally outperformed GPT-4 in precision (97.7% vs 96.8%). GPT-4 showed greater consistency, replicating the exact same results in 76% of the reports across 10 repeated analyses, compared to 59% for GPT-3.5, indicating more stable and reliable performance. Physicians were more likely to miss explicit comorbidities, while the GPT models more frequently inferred nonexplicit comorbidities, sometimes correctly, though this also resulted in more false positives.
Conclusions: This study demonstrates that, with well-designed prompts, the large language models examined can match or even surpass medical specialists in extracting information from complex clinical reports. Their superior efficiency in time and costs, along with easy integration with databases, makes them a valuable tool for large-scale data mining and real-world evidence generation.
Keywords: ChatGPT; EHR; LLMs; data mining; electronic health record; large language models; oncology; radiotherapy.
©Amadeo Jesus Wals Zurita, Hector Miras del Rio, Nerea Ugarte Ruiz de Aguirre, Cristina Nebrera Navarro, Maria Rubio Jimenez, David Muñoz Carmona, Carlos Miguez Sanchez. Originally published in JMIR Medical Informatics (https://medinform.jmir.org), 02.01.2025.
Conflict of interest statement
Conflicts of Interest: None declared.
Figures


References
-
- Liu F, Panagiotakos D. Real-world data: a brief review of the methods, applications, challenges and opportunities. BMC Med Res Methodol. 2022 Nov 05;22(1):287. doi: 10.1186/s12874-022-01768-6. https://bmcmedresmethodol.biomedcentral.com/articles/10.1186/s12874-022-... 10.1186/s12874-022-01768-6 - DOI - DOI - PMC - PubMed
-
- Savova GK, Danciu I, Alamudun F, Miller T, Lin C, Bitterman DS, Tourassi G, Warner JL. Use of Natural Language Processing to Extract Clinical Cancer Phenotypes from Electronic Medical Records. Cancer Res. 2019 Nov 01;79(21):5463–5470. doi: 10.1158/0008-5472.CAN-19-0579. https://europepmc.org/abstract/MED/31395609 0008-5472.CAN-19-0579 - DOI - PMC - PubMed
-
- Adamson B, Waskom M, Blarre A, Kelly J, Krismer K, Nemeth S, Gippetti J, Ritten J, Harrison K, Ho G, Linzmayer R, Bansal T, Wilkinson S, Amster G, Estola E, Benedum CM, Fidyk E, Estévez Melissa, Shapiro W, Cohen AB. Approach to machine learning for extraction of real-world data variables from electronic health records. Front Pharmacol. 2023;14:1180962. doi: 10.3389/fphar.2023.1180962. https://europepmc.org/abstract/MED/37781703 1180962 - DOI - PMC - PubMed
-
- Waskom ML, Tan K, Wiberg H, Cohen AB, Wittmershaus B, Shapiro W. A hybrid approach to scalable real-world data curation by machine learning and human experts. medRxiv. 2023 doi: 10.1101/2023.03.06.23286770. - DOI
MeSH terms
LinkOut - more resources
Full Text Sources

