Multiomic approaches for endotype discovery in allergy/immunology
- PMID: 39746555
- PMCID: PMC12513499
- DOI: 10.1016/j.jaci.2024.12.1083
Multiomic approaches for endotype discovery in allergy/immunology
Keywords: Multiomics; allergy; artificial intelligence; asthma; atopic dermatitis; endotype; epigenome; food allergy; genome; machine learning; metabolome; microbiome; omic; systems biology; transcriptome.
Conflict of interest statement
Disclosure statement Supported by the National Institutes of Health (grants UM1 AI173380, UM AI182034, R01 AI147028, and R01 AI118833). Disclosure of potential conflict of interest: The authors declare that they have no relevant conflicts of interest.
Figures
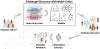

References
-
- Lotvall J, Akdis CA, Bacharier LB, Bjermer L, Casale TB, Custovic A, et al. Asthma endotypes: a new approach to classification of disease entities within the asthma syndrome. J Allergy Clin Immunol 2011; 127:355–60. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

