ECSIT-X4 is Required for Preventing Pressure Overload-Induced Cardiac Hypertrophy via Regulating Mitochondrial STAT3
- PMID: 39746855
- PMCID: PMC11848529
- DOI: 10.1002/advs.202414358
ECSIT-X4 is Required for Preventing Pressure Overload-Induced Cardiac Hypertrophy via Regulating Mitochondrial STAT3
Abstract
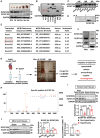
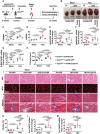
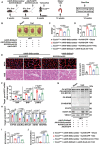
Mitochondrial dysfunction is a key factor in exacerbating pressure overload-induced cardiac hypertrophy and is linked to increased morbidity and mortality. ECSIT, a crucial adaptor for inflammation and mitochondrial function, has been reported to express multiple transcripts in various species and tissues, leading to distinct protein isoforms with diverse subcellular localizations and functions. However, whether an unknown ECSIT isoform exists in cardiac cells and its potential role in regulating mitochondrial function and pathological cardiac hypertrophy has remained unclear. This study identified a 42-kDa ECSIT isoform encoded by the transcript variant Ecsit-X4, which is highly expressed in the mitochondria of adult cardiomyocytes but down-regulated in hypertrophic human heart samples and TAC-treated mouse hearts. AAV9-mediated Ecsit-X4 gene therapy, administered either before or after TAC surgery, significantly attenuated cardiac hypertrophy. Cardiomyocyte-specific Ecsit deficiency worsened TAC-induced cardiac hypertrophy, while Ecsit-X4 compensation independently rescued hypertrophic phenotypes in EcsitcKO mice. Mechanistically, ECSIT-X4 localized to the mitochondria and interacted with STAT3, leading to increased STAT3 levels and enhanced serine 727 phosphorylation in cardiomyocyte mitochondria, thereby promoting strong mitochondrial bioenergetics. This study identified a novel transcript variant of ECSIT localized in the mitochondria of adult cardiomyocytes and highlights its potential as a therapeutic target for heart failure.
Keywords: ECSIT‐X4; STAT3; cardiac hypertrophy; mitochondria.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Nakamura M., Sadoshima J., Nat. Rev. Cardiol. 2018, 15, 387. - PubMed
-
- a) Lopaschuk G. D., Karwi Q. G., Tian R., Wende A. R., Abel E. D., Circ. Res. 2021, 128, 1487; - PMC - PubMed
- b) Kim S. Y., Zhang X., Schiattarella G. G., Altamirano F., Ramos T. A. R., French K. M., Jiang N., Szweda P. A., Evers B. M., May H. I., Luo X., Li H., Szweda L. I., Maracaja‐Coutinho V., Lavandero S., Gillette T. G., Hill J. A., Circulation 2020, 142, 2356. - PMC - PubMed
-
- Liu J., Wang C., Murakami Y., Gong G., Ishibashi Y., Prody C., Ochiai K., Bache R. J., Godinot C., Zhang J., Am. J. Physiol. Heart Circ. Physiol. 2001, 281, H1319. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
