STAT1 regulates immune-mediated intestinal stem cell proliferation and epithelial regeneration
- PMID: 39746933
- PMCID: PMC11697299
- DOI: 10.1038/s41467-024-55227-5
STAT1 regulates immune-mediated intestinal stem cell proliferation and epithelial regeneration
Abstract
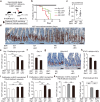
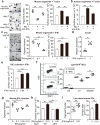
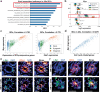
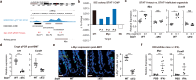
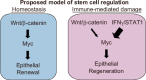
The role of the immune system in regulating tissue stem cells remains poorly understood, as does the relationship between immune-mediated tissue damage and regeneration. Graft vs. host disease (GVHD) occurring after allogeneic bone marrow transplantation (allo-BMT) involves immune-mediated damage to the intestinal epithelium and its stem cell compartment. To assess impacts of T-cell-driven injury on distinct epithelial constituents, we have performed single cell RNA sequencing on intestinal crypts following experimental BMT. Intestinal stem cells (ISCs) from GVHD mice have exhibited global transcriptomic changes associated with a substantial Interferon-γ response and upregulation of STAT1. To determine its role in crypt function, STAT1 has been deleted within murine intestinal epithelium. Following allo-BMT, STAT1 deficiency has resulted in reduced epithelial proliferation and impaired ISC recovery. Similarly, epithelial Interferon-γ receptor deletion has also attenuated proliferation and ISC recovery post-transplant. Investigating the mechanistic basis underlying this epithelial response, ISC STAT1 expression in GVHD has been found to correlate with upregulation of ISC c-Myc. Furthermore, activated T cells have stimulated Interferon-γ-dependent epithelial regeneration in co-cultured organoids, and Interferon-γ has directly induced STAT1-dependent c-Myc expression and ISC proliferation. These findings illustrate immunologic regulation of a core tissue stem cell program after damage and support a role for Interferon-γ as a direct contributor to epithelial regeneration.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing financial interests. A.M.H. and C.A.L hold intellectual property related to Interleukin-22, and A.M.H. has a collaboration with Evive Biotechnology (Shanghai) Ltd, which supported a multicenter clinical trial studying use of Interleukin-22 in patients with GVHD. A.M.H. also serves in a volunteer capacity as a member of the Board of Directors of the American Society for Transplantation and Cellular Therapy (ASTCT).
Figures


References
-
- Sato, T. et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature459, 262–265 (2009). - PubMed
-
- Barker, N. et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature449, 1003–1007 (2007). - PubMed
-
- van der Flier, L. G., Haegebarth, A., Stange, D. E., van de Wetering, M. & Clevers, H. OLFM4 is a robust marker for stem cells in human intestine and marks a subset of colorectal cancer cells. Gastroenterology137, 15–17 (2009). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HL145631/HL/NHLBI NIH HHS/United States
- R01 HL146338/HL/NHLBI NIH HHS/United States
- R01 HL125571/HL/NHLBI NIH HHS/United States
- R01-HL145631/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- R01-HL146338/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

