Liver-specific gene PGRMC1 blocks c-Myc-induced hepatocarcinogenesis through ER stress-independent PERK activation
- PMID: 39747098
- PMCID: PMC11696091
- DOI: 10.1038/s41467-024-55745-2
Liver-specific gene PGRMC1 blocks c-Myc-induced hepatocarcinogenesis through ER stress-independent PERK activation
Abstract
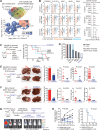
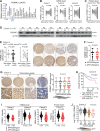
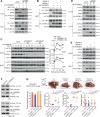
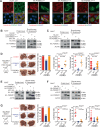
Roles of liver-specific genes (LSGs) in tumor initiation and progression are rarely explored in hepatocellular carcinoma (HCC). Here we show that LSGs are generally downregulated in HCC tumor tissues compared to non-HCC liver tissues, and low-LSG HCCs show poor prognosis and the activated c-Myc pathway. Among the c-Myc- and patient prognosis-associated LSGs, PGRMC1 significantly blocks c-Myc-induced orthotopic HCC formation. The role of PGRMC1 depends on its localization to the endoplasmic reticulum (ER) membrane, where PGRMC1 interacts with PERK through their ER luminal domains. This interaction in turn activates PERK in an ER stress-independent manner, which phosphorylates eIF2α and consequently inhibits c-Myc protein translation. In HCC patients, PGRMC1 level is significantly reduced in tumor tissues and negatively associated with the c-Myc signature. Patients with low-PGRMC1 in their tumors have poor prognosis. Collectively, deregulated LSGs in HCC are associated with the c-Myc pathway activation and PGRMC1 blocks c-Myc-induced hepatic carcinogenesis through promoting ER stress-independent PERK activation.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

