NHSL3 controls single and collective cell migration through two distinct mechanisms
- PMID: 39747206
- PMCID: PMC11696792
- DOI: 10.1038/s41467-024-55647-3
NHSL3 controls single and collective cell migration through two distinct mechanisms
Abstract
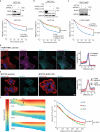
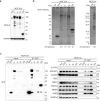
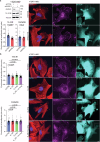
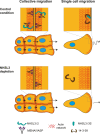
The molecular mechanisms underlying cell migration remain incompletely understood. Here, we show that knock-out cells for NHSL3, the most recently identified member of the Nance-Horan Syndrome family, are more persistent than parental cells in single cell migration, but that, in wound healing, follower cells are impaired in their ability to follow leader cells. The NHSL3 locus encodes several isoforms. We identify the partner repertoire of each isoform using proteomics and predict direct partners and their binding sites using an AlphaFold2-based pipeline. Rescue with specific isoforms, and lack of rescue when relevant binding sites are mutated, establish that the interaction of a long isoform with MENA/VASP proteins is critical at cell-cell junctions for collective migration, while the interaction of a short one with 14-3-3θ in lamellipodia is critical for single cell migration. Taken together, these results demonstrate that NHSL3 regulates single and collective cell migration through distinct mechanisms.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
-
- Ridley, A. J. Life at the leading edge. Cell145, 1012–1022 (2011). - PubMed
-
- Krause, M. & Gautreau, A. Steering cell migration: lamellipodium dynamics and the regulation of directional persistence. Nat. Rev. Mol. Cell Biol.15, 577–590 (2014). - PubMed
-
- Brieher, W. M. & Yap, A. S. Cadherin junctions and their cytoskeleton(s). Curr. Opin. Cell Biol.25, 39–46 (2013). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- ANR-20-CE13-0016/Agence Nationale de la Recherche (French National Research Agency)
- ANR-22-CE13-0041/Agence Nationale de la Recherche (French National Research Agency)
- ANR-21-CE44-0009/Agence Nationale de la Recherche (French National Research Agency)
- ANR-24-CE44-4957/Agence Nationale de la Recherche (French National Research Agency)
- ARC PJA 2021 060003815/Fondation ARC pour la Recherche sur le Cancer (ARC Foundation for Cancer Research)
LinkOut - more resources
Full Text Sources
Research Materials

