PathCrisp: an innovative molecular diagnostic tool for early detection of NDM-resistant infections
- PMID: 39747369
- PMCID: PMC11696219
- DOI: 10.1038/s41598-024-84832-z
PathCrisp: an innovative molecular diagnostic tool for early detection of NDM-resistant infections
Abstract
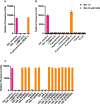
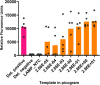
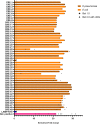
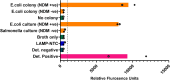
The rapid and early detection of infections and antibiotic resistance markers is a critical challenge in healthcare. Currently, most commercial diagnostic tools for analyzing antimicrobial resistance patterns of pathogens require elaborate culture-based testing. Our study aims to develop a rapid, accurate molecular detection system that can be used directly from culture, thereby introducing molecular testing in conjunction with culture tests to reduce turnaround time and guide therapy. PathCrisp assay, a combination of loop-mediated isothermal amplification and CRISPR-based detection, maintained at a single temperature, was designed and tested on clinical isolates. The specificity and sensitivity of the assay was analyzed, post which the assay was compared with the polymerase chain reaction (PCR) method to detect the New Delhi metallo-beta-lactamase (NDM) gene in carbapenem-resistant enterobacteriaceae clinical samples. Our PathCrisp assay demonstrated the ability to detect as few as 700 copies of the NDM gene from clinical isolates. Our assay demonstrated 100% concordance with the PCR-Sanger sequencing method, more commonly used. Additionally, the lack of the need for a kit-based DNA purification step, rather a crude extraction via heating, enables the direct use of culture samples. The PathCrisp assay is precise, specific and rapid, providing results in approximately 2 h, and operates at a constant temperature, reducing the need for complex equipment handling. In the near future, we hope that this assay can be further optimized and designed as a point-of-care test kit, facilitating its use in various healthcare settings and aiding clinicians in the choice of antibiotics for therapy.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures

Similar articles
-
Evaluation of LAMP assay using phenotypic tests and conventional PCR for detection of blaNDM-1 and blaKPC genes among carbapenem-resistant clinical Gram-negative isolates.J Med Microbiol. 2013 Oct;62(Pt 10):1540-1544. doi: 10.1099/jmm.0.059907-0. Epub 2013 Jun 25. J Med Microbiol. 2013. PMID: 23800599
-
Diagnostic performance of the Xpert Carba-R™ assay directly from rectal swabs for active surveillance of carbapenemase-producing organisms in the largest Brazilian University Hospital.J Microbiol Methods. 2020 Apr;171:105884. doi: 10.1016/j.mimet.2020.105884. Epub 2020 Mar 3. J Microbiol Methods. 2020. PMID: 32142746
-
Real-time PCR assay and a synthetic positive control for the rapid and sensitive detection of the emerging resistance gene New Delhi Metallo-β-lactamase-1 (bla(NDM-1)).Med Microbiol Immunol. 2011 May;200(2):137-41. doi: 10.1007/s00430-011-0189-y. Epub 2011 Feb 25. Med Microbiol Immunol. 2011. PMID: 21350860
-
Rapid detection of carbapenemase-producing Enterobacteriaceae (CPE) using a simple DNA extraction method and Loop-mediated isothermal amplification (LAMP).J Microbiol Methods. 2019 Dec;167:105746. doi: 10.1016/j.mimet.2019.105746. Epub 2019 Nov 5. J Microbiol Methods. 2019. PMID: 31669655 No abstract available.
-
NDM-beta-lactamase-1: Where do we stand?Indian J Med Res. 2022 Feb;155(2):243-252. doi: 10.4103/ijmr.IJMR_685_19. Indian J Med Res. 2022. PMID: 35946201 Free PMC article. Review.
References
-
- WHO. Antibacterial Agents in Clinical and Preclinical Development: An Overview and Analysis (World Health Organization, Geneva, 2024).
-
- Laxminarayan, R. et al. Expanding antibiotic, vaccine, and diagnostics development and access to tackle antimicrobial resistance. Lancet Lond. Engl.403, 2534–2550 (2024). - PubMed
-
- WHO. Guidelines for the Prevention and Control of Carbapenem-Resistant Enterobacteriaceae, Acinetobacter baumannii and Pseudomonas aeruginosa in Health Care Facilities (World Health Organization, Geneva, 2017). - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

