Machine learning-based analysis of microfluidic device immobilized C. elegans for automated developmental toxicity testing
- PMID: 39747450
- PMCID: PMC11696900
- DOI: 10.1038/s41598-024-84842-x
Machine learning-based analysis of microfluidic device immobilized C. elegans for automated developmental toxicity testing
Abstract
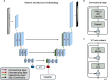
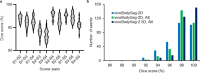
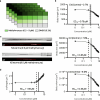
Developmental toxicity (DevTox) tests evaluate the adverse effects of chemical exposures on an organism's development. Although current testing primarily relies on large mammalian models, the emergence of new approach methodologies (NAMs) is encouraging industries and regulatory agencies to evaluate novel assays. C. elegans have emerged as NAMs for rapid toxicity testing because of its biological relevance and suitability to high throughput studies. However, current low-resolution and labor-intensive methodologies prohibit its application for sub-lethal DevTox studies at high throughputs. With the recent advent of the large-scale microfluidic device, vivoChip, we can now rapidly collect 3D high-resolution images of ~ 1000 C. elegans from 24 different populations. While data collection is rapid, analyzing thousands of images remains time-consuming. To address this challenge, we developed a machine-learning (ML)-based image analysis platform using a 2.5D U-Net architecture (vivoBodySeg) that accurately segments C. elegans in images obtained from vivoChip devices, achieving a Dice score of 97.80%. vivoBodySeg processes 36 GB data per device, phenotyping multiple body parameters within 35 min on a desktop PC. This analysis is ~ 140 × faster than the manual analysis. This ML approach delivers highly reproducible DevTox parameters (4-8% CV) to assess the toxicity of chemicals with high statistical power.
Keywords: C. elegans; Developmental toxicity; Few-shot learning; High-throughput screening; Microfluidics; U-Net.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: E.H., S.M., and A.B. are co-founders of vivoVerse, LLC and its Associates. A.D., A.S., A.L., E.H., S.M., and A.B. are inventors of several approved and pending patents. A.M. and S.G. at the time of their contribution are employed by vivoVerse, LLC and have no conflict of interest to declare.
Figures



Update of
-
vivoBodySeg: Machine learning-based analysis of C. elegans immobilized in vivoChip for automated developmental toxicity testing.Res Sq [Preprint]. 2024 Sep 4:rs.3.rs-4796642. doi: 10.21203/rs.3.rs-4796642/v1. Res Sq. 2024. Update in: Sci Rep. 2025 Jan 2;15(1):15. doi: 10.1038/s41598-024-84842-x. PMID: 39281859 Free PMC article. Updated. Preprint.
Similar articles
-
vivoBodySeg: Machine learning-based analysis of C. elegans immobilized in vivoChip for automated developmental toxicity testing.Res Sq [Preprint]. 2024 Sep 4:rs.3.rs-4796642. doi: 10.21203/rs.3.rs-4796642/v1. Res Sq. 2024. Update in: Sci Rep. 2025 Jan 2;15(1):15. doi: 10.1038/s41598-024-84842-x. PMID: 39281859 Free PMC article. Updated. Preprint.
-
Microfluidic systems for high-throughput and high-content screening using the nematode Caenorhabditis elegans.Lab Chip. 2017 Nov 7;17(22):3736-3759. doi: 10.1039/c7lc00509a. Lab Chip. 2017. PMID: 28840220 Review.
-
A novel microfluidic capture and monitoring method for assessing physiological damage of C. elegans under microgravity.Electrophoresis. 2019 Mar;40(6):922-929. doi: 10.1002/elps.201800461. Epub 2019 Jan 7. Electrophoresis. 2019. PMID: 30597589
-
Evaluation of a high-throughput in-vitro-to-in-vivo extrapolation (IVIVE) workflow for the prioritization of potential developmental toxicity of chemicals.ALTEX. 2025 May 28. doi: 10.14573/altex.2406281. Online ahead of print. ALTEX. 2025. PMID: 40434282
-
Microfluidic Technologies for High Throughput Screening Through Sorting and On-Chip Culture of C. elegans.Molecules. 2019 Nov 25;24(23):4292. doi: 10.3390/molecules24234292. Molecules. 2019. PMID: 31775328 Free PMC article. Review.
Cited by
-
Benzo[a]pyrene-Induced Developmental Toxicity in Caenorhabditis elegans: Potential Involvement of Insulin/IGF Signaling and Collagen Gene Dysregulation.Toxics. 2025 May 9;13(5):384. doi: 10.3390/toxics13050384. Toxics. 2025. PMID: 40423463 Free PMC article.
-
AI-CMCA: a deep learning-based segmentation framework for capillary microfluidic chip analysis.Sci Rep. 2025 Jul 21;15(1):26415. doi: 10.1038/s41598-025-11508-7. Sci Rep. 2025. PMID: 40691244 Free PMC article.
References
-
- Kaletta, T. & Hengartner, M. O. Finding function in novel targets: C. elegans as a model organism. Nat. Rev. Drug. Discov.5, 387–398 (2006). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

