Direct specification of lymphatic endothelium from mesenchymal progenitors
- PMID: 39747454
- PMCID: PMC11738995
- DOI: 10.1038/s44161-024-00570-5
Direct specification of lymphatic endothelium from mesenchymal progenitors
Abstract
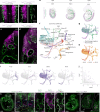
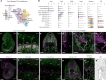
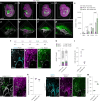
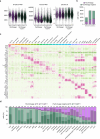
During embryogenesis, endothelial cells (ECs) are generally described to arise from a common pool of progenitors termed angioblasts, which diversify through iterative steps of differentiation to form functionally distinct subtypes of ECs. A key example is the formation of lymphatic ECs (LECs), which are thought to arise largely through transdifferentiation from venous endothelium. Opposing this model, here we show that the initial expansion of mammalian LECs is primarily driven by the in situ differentiation of mesenchymal progenitors and does not require transition through an intermediate venous state. Single-cell genomics and lineage-tracing experiments revealed a population of paraxial mesoderm-derived Etv2+Prox1+ progenitors that directly give rise to LECs. Morphometric analyses of early LEC proliferation and migration, and mutants that disrupt lymphatic development supported these findings. Collectively, this work establishes a cellular blueprint for LEC specification and indicates that discrete pools of mesenchymal progenitors can give rise to specialized subtypes of ECs.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures










References
-
- Huntington, G. S. M. & McClure, C. F. W. The anatomy and development of the jugular lymph sacs in the domestic cat (Felis domestica). Am. J. Anat.10, 177–312 (1910).
MeSH terms
Substances
Grants and funding
- 218561/Z/19/Z/Wellcome Trust (Wellcome)
- 2020-0269/Vetenskapsrådet (Swedish Research Council)
- RE/13/1/30181/BHF Centre of Research Excellence, Oxford (BHF Centre of Research Excellence in Oxford)
- WT_/Wellcome Trust/United Kingdom
- 0011784/Oxford University | John Fell Fund, University of Oxford (John Fell OUP Research Fund)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
