The dynamics of loss of heterozygosity events in genomes
- PMID: 39747660
- PMCID: PMC11811284
- DOI: 10.1038/s44319-024-00353-w
The dynamics of loss of heterozygosity events in genomes
Abstract
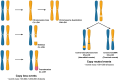
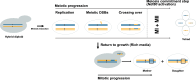
Genomic instability is a hallmark of tumorigenesis, yet it also plays an essential role in evolution. Large-scale population genomics studies have highlighted the importance of loss of heterozygosity (LOH) events, which have long been overlooked in the context of genetic diversity and instability. Among various types of genomic mutations, LOH events are the most common and affect a larger portion of the genome. They typically arise from recombination-mediated repair of double-strand breaks (DSBs) or from lesions that are processed into DSBs. LOH events are critical drivers of genetic diversity, enabling rapid phenotypic variation and contributing to tumorigenesis. Understanding the accumulation of LOH, along with its underlying mechanisms, distribution, and phenotypic consequences, is therefore crucial. In this review, we explore the spectrum of LOH events, their mechanisms, and their impact on fitness and phenotype, drawing insights from Saccharomyces cerevisiae to cancer. We also emphasize the role of LOH in genomic instability, disease, and genome evolution.
Keywords: Saccharomyces cerevisiae; DNA Repair; Genome Instability; Loss of Heterozygosity; Mutation Accumulation.
© 2025. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

