RAGE is a key regulator of ductular reaction-mediated fibrosis during cholestasis
- PMID: 39747668
- PMCID: PMC11811172
- DOI: 10.1038/s44319-024-00356-7
RAGE is a key regulator of ductular reaction-mediated fibrosis during cholestasis
Abstract
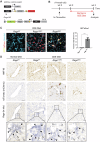
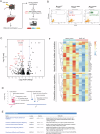
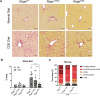
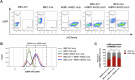
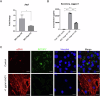
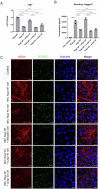
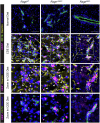
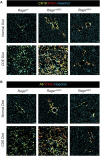
Ductular reaction (DR) is the hallmark of cholestatic diseases manifested in the proliferation of bile ductules lined by biliary epithelial cells (BECs). It is commonly associated with an increased risk of fibrosis and liver failure. The receptor for advanced glycation end products (RAGE) was identified as a critical mediator of DR during chronic injury. Yet, the direct link between RAGE-mediated DR and fibrosis as well as the mode of interaction between BECs and hepatic stellate cells (HSCs) to drive fibrosis remain elusive. Here, we delineate the specific function of RAGE on BECs during DR and its potential association with fibrosis in the context of cholestasis. Employing a biliary lineage tracing cholestatic liver injury mouse model, combined with whole transcriptome sequencing and in vitro analyses, we reveal a role for BEC-specific Rage activity in fostering a pro-fibrotic milieu. RAGE is predominantly expressed in BECs and contributes to DR. Notch ligand Jagged1 is secreted from activated BECs in a Rage-dependent manner and signals HSCs in trans, eventually enhancing fibrosis during cholestasis.
Keywords: Biliary Epithelial Cells; Chronic Liver Injury; Genetically Modified Mice; Hepatic Stellate Cells; Receptor for Advanced Glycation End Products.
© 2024. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. The authors declare no competing interests.
Figures


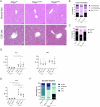
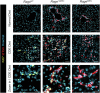


References
-
- Akhurst B, Croager EJ, Farley-Roche CA, Ong JK, Dumble ML, Knight B, Yeoh GC (2001) A modified choline-deficient, ethionine-supplemented diet protocol effectively induces oval cells in mouse liver. Hepatology 34:519–522 - PubMed
-
- Bierhaus A, Humpert PM, Morcos M, Wendt T, Chavakis T, Arnold B, Stern DM, Nawroth PP (2005) Understanding RAGE, the receptor for advanced glycation end products. J Mol Med 83:876–886 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

