Integrated bioinformatics analysis identifies ALDH18A1 as a prognostic hub gene in glutamine metabolism in lung adenocarcinoma
- PMID: 39747729
- PMCID: PMC11695527
- DOI: 10.1007/s12672-024-01698-3
Integrated bioinformatics analysis identifies ALDH18A1 as a prognostic hub gene in glutamine metabolism in lung adenocarcinoma
Abstract
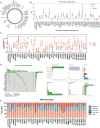
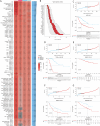
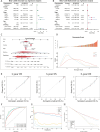
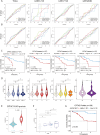
Glutamine metabolism is pivotal in cancer biology, profoundly influencing tumor growth, proliferation, and resistance to therapies. Cancer cells often exhibit an elevated dependence on glutamine for essential functions such as energy production, biosynthesis of macromolecules, and maintenance of redox balance. Moreover, altered glutamine metabolism can contribute to the formation of an immune-suppressive tumor microenvironment characterized by reduced immune cell infiltration and activity. In this study on lung adenocarcinoma, we employed consensus clustering and applied 101 types of machine learning methods to systematically identify key genes associated with glutamine metabolism and develop a risk model. This comprehensive approach provided a clearer understanding of how glutamine metabolism associates with cancer progression and patient outcomes. Notably, we constructed a robust nomogram based on clinical information and patient risk scores, which achieved a stable area under the curve (AUC) greater than 0.8 for predicting patient survival across four datasets, demonstrating high predictive accuracy. This nomogram not only enhances our ability to stratify patient risk but also offers potential targets for therapeutic intervention aimed at disrupting glutamine metabolism and sensitizing tumors to existing treatments. Moreover, we identified ALDH18A1 as a prognostic hub gene of glutamine metabolism, characterized by high expression levels in glutamine cluster 3, which is associated with poor clinical outcomes and worse survival, and is included in the risk model. Such insights underscore the critical role of glutamine metabolism in cancer and highlight avenues for personalized medicine in oncology research.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
