Paracrine role of endothelial IGF-1 receptor in depot-specific adipose tissue adaptation in male mice
- PMID: 39747815
- PMCID: PMC11696296
- DOI: 10.1038/s41467-024-54669-1
Paracrine role of endothelial IGF-1 receptor in depot-specific adipose tissue adaptation in male mice
Abstract
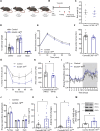
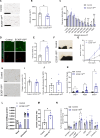
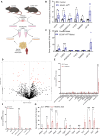
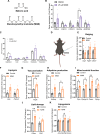
During recent decades, changes in lifestyle have led to widespread nutritional obesity and its related complications. Remodelling adipose tissue as a therapeutic goal for obesity and its complications has attracted much attention and continues to be actively explored. The endothelium lines all blood vessels and is close to all cells, including adipocytes. The endothelium has been suggested to act as a paracrine organ. We explore the role of endothelial insulin-like growth factor-1 receptor (IGF-1R), as a paracrine modulator of white adipose phenotype. We show that a reduction in endothelial IGF-1R expression in the presence of high-fat feeding in male mice leads to depot-specific beneficial white adipose tissue remodelling, increases whole-body energy expenditure and enhances insulin sensitivity via a non-cell-autonomous paracrine mechanism. We demonstrate that increased endothelial malonate may be contributory and that malonate prodrugs have potentially therapeutically relevant properties in the treatment of obesity-related metabolic disease.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

