Development of small molecule non-covalent coronavirus 3CL protease inhibitors from DNA-encoded chemical library screening
- PMID: 39747827
- PMCID: PMC11696555
- DOI: 10.1038/s41467-024-55421-5
Development of small molecule non-covalent coronavirus 3CL protease inhibitors from DNA-encoded chemical library screening
Abstract
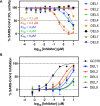
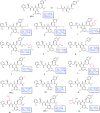
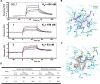
Variants of SARS-CoV-2 have continued to emerge across the world and cause hundreds of deaths each week. Due to the limited efficacy of vaccines against SARS-CoV-2 and resistance to current therapies, additional anti-viral therapeutics with pan-coronavirus activity are of high interest. Here, we screen 2.8 billion compounds from a DNA-encoded chemical library and identify small molecules that are non-covalent inhibitors targeting the conserved 3CL protease of SARS-CoV-2 and other coronaviruses. We perform structure-based optimization, leading to the creation of a series of potent, non-covalent SARS-CoV-2 3CL protease inhibitors, for coronavirus infections. To characterize their binding mechanism to the 3CL protease, we determine 16 co-crystal structures and find that optimized inhibitors specifically interact with both protomers of the native homodimer of 3CL protease. Since 3CL protease is catalytically competent only in the dimeric state, these data provide insight into the design of drug-like inhibitors targeting the native homodimer state. With a binding mode different from the covalent 3CL inhibitor nirmatrelvir, the protease inhibitor in the COVID drug Paxlovid, these compounds may overcome resistance reported for nirmatrelvir and complement its clinical utility.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: S.I., H.L., A.Z., F.F., B.R.S., A.C., and D.D.H. are inventors on a patent application submitted based on this work. B.R.S. is an inventor on additional patents and patent applications related to small molecule therapeutics, and co-founded and serves as a consultant to Exarta Therapeutics, and ProJenX Inc., serves as a consultant to Weatherwax Biotechnologies and Akin Gump Strauss Hauer & Feld LLP, and holds equity in Sonata Therapeutics. S.I. and D.D.H. have sponsored research agreements with Enanta Pharmaceuticals, Shionogi & Co., Ltd., and Regeneron Pharmaceuticals. D.D.H. is a co-founder of TaiMed Biologics and RenBio, consultant to WuXi Biologics, Brii Biosciences, Apexigen, and Veru Inc., and board director for Vicarious Surgical. The remaining authors declare no competing interests.
Figures






References
-
- WHO. WHO Coronavirus (COVID-19) Dashboard, https://covid19.who.int (2023).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

