Molecular insights into a distinct class of terpenoid cyclases
- PMID: 39747870
- PMCID: PMC11695735
- DOI: 10.1038/s41467-024-55717-6
Molecular insights into a distinct class of terpenoid cyclases
Abstract
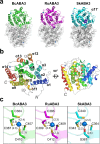
Terpenoid cyclases (TCs) account for the synthesis of the most widespread and diverse natural compounds. A sesquiterpene cyclase termed BcABA3 from an abscisic acid-producing fungus Botrytis cinerea that yields (2Z,4E)-α-ionylideneethane but lacks signature feature of canonical TCs represents a distinct type of TCs. Here, we report the crystal structures of BcABA3, a closely related RuABA3 from Rutstroemia sp. and a bacterial SkABA3 from Shimazuella kribbensis. These ABA3 proteins adopt an all-α-helix fold and bind pyrophosphate moiety of farnesyl pyrophosphate by Glu-chelated Mg2+ ion cluster. We conduct mutagenesis experiments to validate the role of the substrate-binding residues. SkABA3 appears to yield compounds that are distinct from (2Z,4E)-α-ionylideneethane. These results not only provide the molecular insight into ABA3 proteins that serve as an important basis to the future investigation of this class of TCs, but also reveal the existence of more uncharacterized terpenoids synthesized via dedicated machineries.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
-
- Yang, W. et al. Advances in pharmacological activities of terpenoids. Nat. Prod. Commun.15, 1–13 (2020).
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

