L3MBTL3 and STAT3 collaboratively upregulate SNAIL expression to promote metastasis in female breast cancer
- PMID: 39747894
- PMCID: PMC11696420
- DOI: 10.1038/s41467-024-55617-9
L3MBTL3 and STAT3 collaboratively upregulate SNAIL expression to promote metastasis in female breast cancer
Abstract
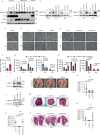
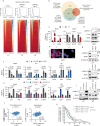
The STAT3 pathway promotes epithelial-mesenchymal transition, migration, invasion and metastasis in cancer. STAT3 upregulates the transcription of the key epithelial-mesenchymal transition transcription factor SNAIL in a DNA binding-independent manner. However, the mechanism by which STAT3 is recruited to the SNAIL promoter to upregulate its expression is still elusive. In our study, the lysine methylation binding protein L3MBTL3 is positively associated with metastasis and poor prognosis in female patients with breast cancer. L3MBTL3 also promotes epithelial-mesenchymal transition and metastasis in breast cancer. Mechanistic analysis reveals that L3MBTL3 interacts with STAT3 and recruits STAT3 to the SNAIL promoter to increase SNAIL transcription levels. The interaction between L3MBTL3 and STAT3 is required for SNAIL transcription upregulation and metastasis in breast cancer, while the methylated lysine binding activity of L3MBTL3 is not required for these functions. In conclusion, L3MBTL3 and STAT3 synergistically upregulate SNAIL expression to promote breast cancer metastasis.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures








References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

