Recognition of Self and Viral Ligands by NK Cell Receptors
- PMID: 39748148
- PMCID: PMC11695704
- DOI: 10.1111/imr.13435
Recognition of Self and Viral Ligands by NK Cell Receptors
Abstract
Natural killer (NK) cells are essential elements of the innate immune response against tumors and viral infections. NK cell activation is governed by NK cell receptors that recognize both cellular (self) and viral (non-self) ligands, including MHC, MHC-related, and non-MHC molecules. These diverse receptors belong to two distinct structural families, the C-type lectin superfamily and the immunoglobulin superfamily. NK receptors include Ly49s, KIRs, LILRs, and NKG2A/CD94, which bind MHC class I (MHC-I) molecules, and NKG2D, which binds MHC-I paralogs such MICA and ULBP. Other NK receptors recognize tumor-associated antigens (NKp30, NKp44, NKp46), cell-cell adhesion proteins (KLRG1, CD96), or genetically coupled C-type lectin-like ligands (NKp65, NKR-P1). Additionally, cytomegaloviruses have evolved various immunoevasins, such as m157, m12, and UL18, which bind NK receptors and act as decoys to enable virus-infected cells to escape NK cell-mediated lysis. We review the remarkable progress made in the past 25 years in determining structures of representatives of most known NK receptors bound to MHC, MHC-like, and non-MHC ligands. Together, these structures reveal the multiplicity of solutions NK receptors have developed to recognize these molecules, and thereby mediate crucial interactions for regulating NK cytolytic activity by self and viral ligands.
Keywords: KIR; Ly49; MHC; NK receptor; structure; virus.
© 2025 The Author(s). Immunological Reviews published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

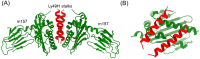




References
-
- Di Santo J. P., “Natural Killer Cells: Diversity in Search of a Niche,” Nature Immunology 9 (2008): 473–475. - PubMed
-
- Vivier E., Tomasello E., Baratin M., Walzer T., and Ugolini S., “Functions of Natural Killer Cells,” Nature Immunology 9 (2008): 503–510. - PubMed
-
- Cerwenka A. and Lanier L. L., “Natural Killer Cell Memory in Infection, Inflammation and Cancer,” Nature Reviews. Immunology 16 (2016): 112–123. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

