A new peptide inhibitor of C1QBP exhibits potent anti-tumour activity against triple negative breast cancer by impairing mitochondrial function and suppressing homologous recombination repair
- PMID: 39748215
- PMCID: PMC11695203
- DOI: 10.1002/ctm2.70162
A new peptide inhibitor of C1QBP exhibits potent anti-tumour activity against triple negative breast cancer by impairing mitochondrial function and suppressing homologous recombination repair
Abstract
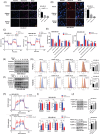
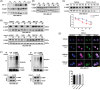
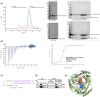
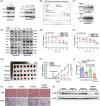
C1QBP exhibits heightened expression across a spectrum of tumours, thereby fostering their proliferation and metastasis, rendering it a pivotal therapeutic target. Nevertheless, to date, no pharmacological agents capable of directly targeting and inducing the degradation of C1QBP have been identified. In this study, we have unveiled a new peptide, PDBAG1, derived from the precursor protein GPD1, employing a peptidomics-based drug screening strategy. PDBAG1 has demonstrated substantial efficacy in suppressing triple-negative breast cancer (TNBC) both in vitro and in vivo. Its mechanism of action involves mitochondrial impairment and the inhibition of oxidative phosphorylation (OXPHOS), achieved through direct binding to C1QBP, thereby promoting its ubiquitin-dependent degradation. Concomitantly, due to metabolic adaptability, we have observed an up-regulation of glycolysis to compensate for OXPHOS inhibition. We observed an aberrant phenomenon wherein the hypoxia signalling pathway in tumour cells exhibited significant activation under normoxic conditions following PDBAG1 treatment. Through size-exclusion chromatography (SEC) and isothermal titration calorimetry (ITC) assays, we have validated that PDBAG1 is capable of binding C1QBP with a Kd value of 334 nM. Furthermore, PDBAG1 inhibits homologous recombination repair proteins and facilitates synergism with poly-ADP-ribose polymerase inhibitors in cancer therapy. This underscores that PDBAG1 ultimately induces insurmountable survival stress through multiple mechanisms while concurrently engendering therapeutic vulnerabilities specific to TNBC. KEY POINTS: The newly discovered peptide PDBAG1 is the first small molecule substance found to directly target and degrade C1QBP, demonstrating significant tumour inhibitory effects and therapeutic potential.
Keywords: C1QBP; anti‐tumour peptide; homologous recombination repair; mitochondrial function; targeted protein degradation.
© 2025 The Author(s). Clinical and Translational Medicine published by John Wiley & Sons Australia, Ltd on behalf of Shanghai Institute of Clinical Bioinformatics.
Conflict of interest statement
The authors have no conflicts of interest to declare.
Figures



Similar articles
-
The cancer-testis gene, MEIOB, sensitizes triple-negative breast cancer to PARP1 inhibitors by inducing homologous recombination deficiency.Cancer Biol Med. 2021 Feb 15;18(1):74-87. doi: 10.20892/j.issn.2095-3941.2020.0071. Cancer Biol Med. 2021. PMID: 33628586 Free PMC article.
-
mTOR Inhibitors Suppress Homologous Recombination Repair and Synergize with PARP Inhibitors via Regulating SUV39H1 in BRCA-Proficient Triple-Negative Breast Cancer.Clin Cancer Res. 2016 Apr 1;22(7):1699-712. doi: 10.1158/1078-0432.CCR-15-1772. Epub 2015 Nov 6. Clin Cancer Res. 2016. PMID: 26546619 Free PMC article.
-
A novel UCHL3 inhibitor, perifosine, enhances PARP inhibitor cytotoxicity through inhibition of homologous recombination-mediated DNA double strand break repair.Cell Death Dis. 2019 May 21;10(6):398. doi: 10.1038/s41419-019-1628-8. Cell Death Dis. 2019. PMID: 31113933 Free PMC article.
-
Homologous recombination deficiency and host anti-tumor immunity in triple-negative breast cancer.Breast Cancer Res Treat. 2018 Aug;171(1):21-31. doi: 10.1007/s10549-018-4807-x. Epub 2018 May 7. Breast Cancer Res Treat. 2018. PMID: 29736741 Review.
-
Poly (adenosine diphosphate-ribose) polymerase inhibitors in the treatment of triple-negative breast cancer with homologous repair deficiency.Med Res Rev. 2024 Nov;44(6):2774-2792. doi: 10.1002/med.22058. Epub 2024 Jun 24. Med Res Rev. 2024. PMID: 38922930 Review.
References
MeSH terms
Substances
Grants and funding
- 82102739/National Natural Science Foundation of China
- 81502307/National Natural Science Foundation of China
- JQX21009/Nanjing Medical Science and Technique Development Foundation
- YKK21163/Nanjing Medical Science and Technique Development Foundation
- YKK20139/Nanjing Medical Science and Technique Development Foundation
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
