Role of the Notch signaling pathway in porcine oocyte maturation
- PMID: 39748238
- PMCID: PMC11697911
- DOI: 10.1186/s12964-024-01996-x
Role of the Notch signaling pathway in porcine oocyte maturation
Abstract
Background: Although the Notch signaling pathway is known to play an important role in ovarian follicle development in mammals, whether it is involved in oocyte maturation remains unclear. Therefore, this study was performed to elucidate the existence and role of the Notch signaling pathway during oocyte maturation in a porcine model.
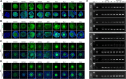
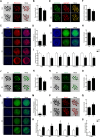
Methods: Reverse transcription-polymerase chain reaction (RT-PCR) and immunocytochemical assays were used to determine the existence of Notch signaling pathway-related transcripts and proteins in porcine cumulus-oocyte complexes (COCs). In vitro maturation (IVM) and parthenogenetic activation of oocytes were employed to examine the effects of Notch signaling inhibition on meiotic progression and embryogenesis of COCs using RO4929097 (RO), an inhibitor of γ secretase. Various staining methods (TUNEL, Phalloidin-TRITC, MitoTracker, JC-1, BODIPY FL ATP, ER-Tracker, Fluo-3, and Rhod-2) and immunocytochemical and quantitative PCR assays were used to identify the effects of Notch signaling inhibition on meiotic progression, embryogenesis, cell cycle progression, spindle assembly, chromosome alignment, mitochondrial and endoplasmic reticulum distribution, and downstream pathway targets in COCs.
Results: The RT-PCR and immunocytochemical analyses revealed the presence of Notch signaling-related receptors (NOTCH1-4) and ligands (JAG1 and 2 and DLL1, 3, and 4) at 0, 22, 28, and 44 h of IVM in the COCs. RO treatment during oocyte maturation markedly reduced meiotic maturation and embryogenesis, inhibiting the cell cycle progression, spindle assembly, and chromosome alignment processes that are important for meiotic maturation. Furthermore, RO significantly impaired the cellular distribution and functions of the mitochondria and endoplasmic reticula, which are important organelles for the cytoplasmic maturation of oocytes. Finally, the involvement of canonical Notch signaling in oocyte maturation was confirmed by the decreased expression of HES and HEY family transcripts and proteins in the RO-treated COCs.
Conclusions: It was first demonstrated that Notch signaling pathway-related transcripts and proteins were expressed during the meiotic maturation of porcine COCs. Furthermore, the inhibition of Notch signaling during IVM revealed the essential role of this signaling pathway during oocyte maturation in pigs.
Keywords: Embryo development; Microtubule function; Notch signaling pathway; Oocyte maturation; Organelle function; Pig; γ secretase inhibitor.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Animal experiments were approved and reviewed by the Korea Research Institute of Bioscience and Biotechnology (KRIBB) Institutional Animal Care and Use Committee (approval no. KRIBB-AEC-21107, date 03.24.2021) and all studies were conducted in accordance with the Basel Declaration. Consent for publication: We also confirm that all the listed authors have participated actively in the study, and have seen and approved the submitted manuscript. Competing interests: The authors declare no competing interests.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

