Spatiotemporal dynamics of early oogenesis in pigs
- PMID: 39748324
- PMCID: PMC11694410
- DOI: 10.1186/s13059-024-03464-8
Spatiotemporal dynamics of early oogenesis in pigs
Abstract
Background: In humans and other mammals, the process of oogenesis initiates asynchronously in specific ovarian regions, leading to the localization of dormant and growing follicles in the cortex and medulla, respectively; however, the current understanding of this process remains insufficient.
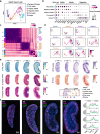
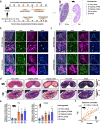
Results: Here, we integrate single-cell RNA sequencing (scRNA-seq) and spatial transcriptomics (ST) to comprehend spatial-temporal gene expression profiles and explore the spatial organization of ovarian microenvironments during early oogenesis in pigs. Projection of the germ cell clusters at different stages of oogenesis into the spatial atlas unveils a "cortical to medullary (C-M)" distribution of germ cells in the developing porcine ovaries. Cross-species analysis between pigs and humans unveils a conserved C-M distribution pattern of germ cells during oogenesis, highlighting the utility of pigs as valuable models for studying human oogenesis in a spatial context. RNA velocity analysis with ST identifies the molecular characteristics and spatial dynamics of granulosa cell lineages originating from the cortical and medullary regions in pig ovaries. Spatial co-occurrence analysis and intercellular communication analysis unveils a distinct cell-cell communication pattern between germ cells and somatic cells in the cortex and medulla regions. Notably, in vitro culture of ovarian tissues verifies that intercellular NOTCH signaling and extracellular matrix (ECM) proteins played crucial roles in initiating meiotic and oogenic programs, highlighting an underappreciated role of ovarian microenvironments in orchestrating germ cell fates.
Conclusions: Overall, our work provides insight into the spatial characteristics of early oogenesis and the regulatory role of ovarian microenvironments in germ cell fate within a spatial context.
Keywords: Early oogenesis; Microenvironments; Spatiotemporal transcriptomics.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: All experimental procedures involving animal experiments were approved by the Ethics Committee of Qingdao Agricultural University (No. SYXK-20220–021). Consent for publication: Not applicable. Competing interests: The authors declare that they have no competing interests.
Figures





References
MeSH terms
LinkOut - more resources
Full Text Sources

