Characterization of the E26H Mutant Schistosoma japonicum Glutathione S-Transferase
- PMID: 39748618
- PMCID: PMC11968563
- DOI: 10.1002/prot.26794
Characterization of the E26H Mutant Schistosoma japonicum Glutathione S-Transferase
Abstract
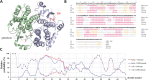
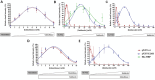
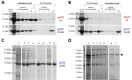
Glutathione-S-transferase, such as that of Schistosoma japonicum (sjGST) belongs to the most widely utilized fusion tags in the recombinant protein technology. The E26H mutation of sjGST has already been found to remarkably improve its ability for binding divalent ions, enabling its purification with immobilized metal affinity chromatography (IMAC). Nevertheless, most characteristics of this mutant remained unexplored to date. In this study, we performed a comparative analysis of the wild-type and the E26H mutant sjGST by using in vitro as well as in silico approaches. We confirmed that the sjGST(E26H) protein exhibits significantly increased affinity for binding nickel ions as compared to the wild-type. In addition, we proved that the sjGST(E26H) can be purified efficiently either with glutathione- or immobilized metal ion-affinity chromatography, even in consecutive purification steps. The human retroviral-like aspartic protease 1 (ASPRV1) conjugated with the sjGST(E26H) fusion tag was also successfully purified by using both of these affinity chromatographic approaches. Our studies revealed that the E26H mutant sjGST can be used as a versatile affinity tag because the modified protein retains the kinetic features of the wild-type and its affinity towards glutathione, while can be purified efficiently by IMAC, as well.
Keywords: Schistosoma japonicum; GST; affinity chromatography; fusion tag; glutathione S‐transferase; protein purification; recombinant protein.
© 2025 The Author(s). PROTEINS: Structure, Function, and Bioinformatics published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
A histidine substitution confers metal binding affinity to a Schistosoma japonicum Glutathione S-transferase.Protein Expr Purif. 2010 Sep;73(1):74-7. doi: 10.1016/j.pep.2010.03.014. Epub 2010 Mar 27. Protein Expr Purif. 2010. PMID: 20347989
-
Structure-based design, biophysical characterization, and biochemical application of the heterodimeric affinity purification tag based on the Schistosoma japonicum glutathione-S-transferase (SjGST) homodimer.J Biochem. 2024 Jul 1;176(1):69-80. doi: 10.1093/jb/mvae028. J Biochem. 2024. PMID: 38471515
-
Affinity purification of Schistosoma japonicum glutathione-S-transferase and its site-directed mutants with glutathione affinity chromatography and immobilized metal affinity chromatography.J Chromatogr A. 1999 Aug 6;852(1):151-9. doi: 10.1016/s0021-9673(99)00490-2. J Chromatogr A. 1999. PMID: 10480240
-
Crystal structure of non-fused glutathione S-transferase from Schistosoma japonicum in complex with glutathione.Protein Pept Lett. 2005 Oct;12(7):709-12. doi: 10.2174/0929866054696154. Protein Pept Lett. 2005. PMID: 16522189
-
Affinity Tags for Protein Purification.Curr Protein Pept Sci. 2020;21(8):821-830. doi: 10.2174/1389203721666200606220109. Curr Protein Pept Sci. 2020. PMID: 32504500 Review.
References
-
- Terpe K., “Overview of Tag Protein Fusions, From Molecular and Biochemical Fundamentals to Commercial Systems,” Applied Microbiology and Biotechnology 60, no. 5 (2003): 523–533. - PubMed
-
- Schäfer F., Seip N., Maertens B., Block H., and Kubicek J., “Purification of GST‐Tagged Proteins,” Methods in Enzymology 559 (2015): 127–139. - PubMed
MeSH terms
Substances
Grants and funding
- TKP2021-EGA-20/National Research, Development, and Innovation Fund (Ministry of Culture and Innovation of Hungary), Thematic Excellence Program (Biotechnology)
- ÚNKP-22-2-I-DE-391/National Research, Development and Innovation Fund (Ministry of Culture and Innovation of Hungary), New National Excellence Program
- ÚNKP-23-5-DE-486/National Research, Development and Innovation Fund (Ministry of Culture and Innovation of Hungary), New National Excellence Program
- BO/00110/23/5/János Bolyai Research Scholarship (Hungarian Academy of Sciences)
- University of Debrecen (UD) Program for Scientific Publication
LinkOut - more resources
Full Text Sources
Research Materials

