Revisiting the emerging pathosystem of rice sheath blight: deciphering the Rhizoctonia solani virulence, host range, and rice genotype-based resistance
- PMID: 39748817
- PMCID: PMC11693681
- DOI: 10.3389/fpls.2024.1499785
Revisiting the emerging pathosystem of rice sheath blight: deciphering the Rhizoctonia solani virulence, host range, and rice genotype-based resistance
Abstract
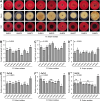
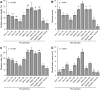
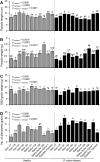
Sheath blight, caused by Rhizoctonia solani AG1 IA, is a challenging disease of rice worldwide. In the current study, nine R. solani isolates, within the anastomosis group AG-1 IA, were isolated, characterized based on their macroscopic and microscopic features, as well as their ability to produce cell wall degrading enzymes (CWDEs), and further molecularly identified via ITS sequencing. Although all isolates were pathogenic and produced typical sheath blight symptoms the susceptible rice cultivar, Sakha 101, R. solani AG1 IA -isolate SHBP9 was the most aggressive isolate. The virulence of isolate SHBP9 was correlated with its overproduction of CWDEs, where it had the highest pectinase, amylase, and cellulase activity in vitro. R. solani AG1 IA -isolate SHBP9 was able to infect 12 common rice-associated weeds from the family Poaceae, as well as over 25 economic crops from different families, except chickpea (Cicer arietinum) from Fabaceae, Rocket (Eruca sativa) from Brassicaceae, and the four crops from Solanaceae. Additionally, rice genotype-based resistance was evaluated using 11 rice genotypes for their response to R. solani isolates, morphological traits, yield components, and using 12 SSR markers linked to sheath blight resistance. Briefly, the tested 11 rice genotypes were divided into three groups; Cluster "I" included only two resistant genotypes (Egyptian Yasmine and Giza 182), Cluster "II" included four moderately resistant genotypes (Egyptian hybrid 1, Giza 178, 181, and 183), whereas Cluster "III" included five susceptible (Sakha 104, 101, 108, Super 300 and Giza 177). Correspondingly, only surface-mycelium growth was microscopically noticed on the resistant cultivar Egyptian Yasmine, as well as the moderately resistant Egyptian hybrid 1, however, on the susceptible Sakha 104, the observed mycelium was branched, shrunk, and formed sclerotia. Accordingly, Indica and Indica/Japonica rice genotypes showed more resistance to R. solani than Japonica genotypes. These findings provide insights into its pathogenicity mechanisms and identify potential targets for disease control which ultimately contributes to the development of sustainable eco-friendly disease management strategies. Moreover, our findings might pave the way for developing resistant rice varieties by using more reliable resistance sources of non-host plants, as well as, rice genotype-based resistance as a genetic resource.
Keywords: SSR marker; cell wall; host range; rhizoctonia; rice genotype; sheath blight; weeds.
Copyright © 2024 Kalboush, Mazrou, Hassan, Sherif, Gabr, Ali and Nehela.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures








References
-
- Ahvenniemi P., Wolf M., Lehtonen M. J., Wilson P., German-Kinnari M., Valkonen J. P. (2009). Evolutionary diversification indicated by compensatory base changes in ITS2 secondary structures in a complex fungal species, Rhizoctonia solani . J. Mol. Evol. 69, 150–163. doi: 10.1007/s00239-009-9260-3 - DOI - PubMed
-
- Ajayi-Oyetunde O. O., Bradley C. A. (2018). Rhizoctonia solani: taxonomy, population biology and management of Rhizoctonia seedling disease of soybean. Plant Pathol. 67, 3–17. doi: 10.1111/ppa.12733 - DOI
-
- Anand P., Alwin A., Vennison S. J., Sankar S. G., Gilwax Prabhu D. I., Vasan P. T., et al. (2010). Isolation and characterization of bacteria from the gut of Bombyx mori that degrade cellulose, xylan, pectin and starch and their impact on digestion. J. Insect Sci. 10, 107. doi: 10.1673/031.010.10701 - DOI - PMC - PubMed
-
- Atallah O. O., Hassanin A., Yassin S. M., Aloufi A. S., Almanzalawi E. A., Abdelkhalek A., et al. (2024). Pathological characterization and management of Lasiodiplodia theobromae, a hemi-biotroph with an interkingdom host range. Plant Dis. 108, 3243–3257. doi: 10.1094/PDIS-03-24-0713-RE - DOI - PubMed
LinkOut - more resources
Full Text Sources

