Computational design of CDK1 inhibitors with enhanced target affinity and drug-likeness using deep-learning framework
- PMID: 39748968
- PMCID: PMC11693894
- DOI: 10.1016/j.heliyon.2024.e40345
Computational design of CDK1 inhibitors with enhanced target affinity and drug-likeness using deep-learning framework
Abstract
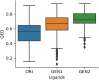
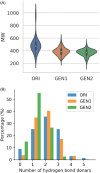
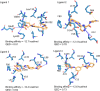
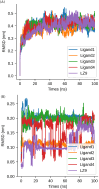
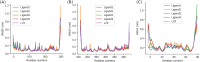
Cyclin Dependent Kinase 1 (CDK1) plays a crucial role in cell cycle regulation, and dysregulation of its activity has been implicated in various cancers. Although several CDK1 inhibitors are currently in clinical trials, none have yet been approved for therapeutic use. This research utilized deep learning techniques, specifically Recurrent Neural Networks with Long Short-Term Memory (LSTM), to generate potential CDK1 inhibitors. Molecular docking, evaluation of molecular properties, and molecular dynamics simulations were conducted to identify the most promising candidates. The results showed that the generated ligands exhibited substantial improvements in target affinity and drug-likeness. Molecular docking results showed that the generated ligands had an average binding affinity of -10.65 ± 0.877 kcal/mol towards CDK1. The Quantitative Estimate of Drug-likeness (QED) values for the generated ligands averaged 0.733 ± 0.10, significantly higher than the 0.547 ± 0.15 observed for known CDK1 inhibitors (p < 0.001). Molecular dynamics simulations further confirmed the stability and favorable interactions of the selected ligands with the CDK1 complex. The identification of novel CDK1 inhibitors with improved binding affinities and drug-likeness properties could potentially fill the gap in the ongoing development of CDK inhibitors. However, it is imperative to note that extensive experimental validation is required prior to advancing these generated ligands to subsequent stages of drug development.
Keywords: CDK1 inhibitors; Cyclin dependent kinase 1 (CDK1); MD simulations; Molecular docking; Recurrent neural network.
© 2024 Published by Elsevier Ltd.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures






References
-
- Finn R.S., Martin M., Rugo H.S., Jones S., Im S., Gelmon K., Harbeck N., Lipatov O.N., Walshe J.M., Moulder S., Gauthier E., Lu D.R., Randolph S., Diéras V., Slamon D.J. Palbociclib and letrozole in advanced breast cancer. N. Engl. J. Med. 2016;375:1925–1936. doi: 10.1056/NEJMoa1607303. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

