Gut microbiota mediated T cells regulation and autoimmune diseases
- PMID: 39749132
- PMCID: PMC11694513
- DOI: 10.3389/fmicb.2024.1477187
Gut microbiota mediated T cells regulation and autoimmune diseases
Abstract
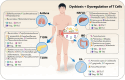
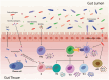
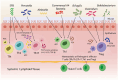
Gut microbiota regulates the immune system, the development and progression of autoimmune diseases (AIDs) and overall health. Recent studies have played a crucial part in understanding the specific role of different gut bacterial strains and their metabolites in different AIDs. Microbial signatures in AIDs are revealed by advanced sequencing and metabolomics studies. Microbes such as Faecalibacterium prausnitzii, Akkermansia muciniphila, Anaerostipes caccae, Bacteroides sp., Roseburia sp., Blautia sp., Blautia faecis, Clostridium lavalense, Christensenellaceae sp., Coprococcus sp., Firmicutes sp., Ruminococcaceae sp., Lachnospiraceae sp., Megamonas sp., Monoglobus sp., Streptococcus pneumoniae and Bifidobacterium sp. help maintain immune homeostasis; whereas, Prevotella copri, Ruminococcus gnavus, Lactobacillus salivarius, Enterococcus gallinarum, Elizabeth menigoseptica, Collinsella sp., Escherichia sp., Fusobacterium sp., Enterobacter ludwigii, Enterobacteriaceae sp., Proteobacteria, Porphyromonas gingivalis, Porphyromonas nigrescens, Dorea sp., and Clostridium sp. cause immuno-pathogenesis. A complex web of interactions is revealed by understanding the influence of gut microbiota on immune cells and various T cell subsets such as CD4+ T cells, CD8+ T cells, natural killer T cells, γδ T cells, etc. Certain AIDs, including rheumatoid arthritis, diabetes mellitus, atopic asthma, inflammatory bowel disease and non-alcoholic fatty liver disease exhibit a state of dysbiosis, characterized by alterations in microbial diversity and relative abundance of specific taxa. This review summarizes recent developments in understanding the role of certain microbiota composition in specific AIDs, and the factors affecting specific regulatory T cells through certain microbial metabolites and also focuses the potential application and therapeutic significance of gut microbiota-based interventions as novel adjunctive therapies for AIDs. Further research to determine the precise association of each gut bacterial strain in specific diseases is required.
Keywords: Anaerostipes caccae; Bacteroides sp.; Blautia faecis; Blautia sp.; Christensenellaceae sp.; Clostridium lavalense; Roseburia sp.; muciniphila.
Copyright © 2024 Bhutta, Xu, Jian, Wang, Liu, Sun, Han, Wu and Javeed.
Conflict of interest statement
YL and SW were employed by the Hangzhou Zheda Dixun Biological Gene Engineering Co., Ltd. The authors declare that this study received funding from Hangzhou Zheda Dixun Biological Gene Engineering Co, Ltd., Anti-Allergy Functional Molecular Laboratory. The funder had the following involvement in the study: reviewing and editing of the manuscript.
Figures
Similar articles
-
Understanding the Role of the Gut Microbiome and Microbial Metabolites in Non-Alcoholic Fatty Liver Disease: Current Evidence and Perspectives.Biomolecules. 2021 Dec 31;12(1):56. doi: 10.3390/biom12010056. Biomolecules. 2021. PMID: 35053205 Free PMC article. Review.
-
Gut Microbiota and Its Repercussion in Parkinson's Disease: A Systematic Review in Occidental Patients.Neurol Int. 2023 Jun 13;15(2):750-763. doi: 10.3390/neurolint15020047. Neurol Int. 2023. PMID: 37368331 Free PMC article. Review.
-
The Relationship Between Pediatric Gut Microbiota and SARS-CoV-2 Infection.Front Cell Infect Microbiol. 2022 Jul 8;12:908492. doi: 10.3389/fcimb.2022.908492. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35873161 Free PMC article.
-
Gallstone Disease, Obesity and the Firmicutes/Bacteroidetes Ratio as a Possible Biomarker of Gut Dysbiosis.J Pers Med. 2020 Dec 25;11(1):13. doi: 10.3390/jpm11010013. J Pers Med. 2020. PMID: 33375615 Free PMC article. Review.
-
The Gut Microbiota of Healthy Chilean Subjects Reveals a High Abundance of the Phylum Verrucomicrobia.Front Microbiol. 2017 Jun 30;8:1221. doi: 10.3389/fmicb.2017.01221. eCollection 2017. Front Microbiol. 2017. PMID: 28713349 Free PMC article.
Cited by
-
Characterization of Gut Microbiota in Patients with Active Spreading Vitiligo Based on Whole-Genome Shotgun Sequencing.Int J Mol Sci. 2025 Mar 24;26(7):2939. doi: 10.3390/ijms26072939. Int J Mol Sci. 2025. PMID: 40243573 Free PMC article.
-
Recent insights and advances in gut microbiota's influence on host antiviral immunity.Front Microbiol. 2025 Feb 27;16:1536778. doi: 10.3389/fmicb.2025.1536778. eCollection 2025. Front Microbiol. 2025. PMID: 40083779 Free PMC article. Review.
-
Interacting roles of gut microbiota and T cells in the development of autoimmune hepatitis.Front Immunol. 2025 May 26;16:1584001. doi: 10.3389/fimmu.2025.1584001. eCollection 2025. Front Immunol. 2025. PMID: 40491914 Free PMC article. Review.
-
Exploring the gut microbiome's influence on cancer-associated anemia: Mechanisms, clinical challenges, and innovative therapies.World J Gastrointest Pharmacol Ther. 2025 Jun 5;16(2):105375. doi: 10.4292/wjgpt.v16.i2.105375. World J Gastrointest Pharmacol Ther. 2025. PMID: 40575364 Free PMC article.
-
The gut-immune axis in primary immune thrombocytopenia (ITP): a paradigm shifts in treatment approaches.Front Immunol. 2025 Jun 12;16:1595977. doi: 10.3389/fimmu.2025.1595977. eCollection 2025. Front Immunol. 2025. PMID: 40574831 Free PMC article. Review.
References
Publication types
LinkOut - more resources
Full Text Sources
Research Materials

