Circulating biomarkers associated with pediatric sickle cell disease
- PMID: 39749215
- PMCID: PMC11694143
- DOI: 10.3389/fmolb.2024.1481441
Circulating biomarkers associated with pediatric sickle cell disease
Abstract
Introduction: Sickle cell disease (SCD) is a genetic blood disorder caused by a mutation in the HBB gene, which encodes the beta-globin subunit of hemoglobin. This mutation leads to the production of abnormal hemoglobin S (HbS), causing red blood cells to deform into a sickle shape. These deformed cells can block blood flow, leading to complications like chronic hemolysis, anemia, severe pain episodes, and organ damage. SCD genotypes include HbSS, HbSC (HbC is an abnormal variant of hemoglobin), and HbS/β-thalassemia. Sickle cell trait (SCT), HbAS, represents the carrier state, while other hemoglobin variants include HbCC, HbAC, and the normal HbAA. Over 7.5 million people worldwide live with SCD, with a high mortality rate in sub-Saharan Africa, including Ghana. Despite its prevalence, SCD is underdiagnosed and poorly managed, especially in children. Characterized by intravascular hemolysis, SCD leads to oxidative stress, endothelial activation, and systemic inflammation. Identifying circulating blood biomarkers indicative of organ damage and systemic processes is vital for understanding SCD and improving patient management. However, research on biomarkers in pediatric SCD is limited and few have been identified and validated. This study explores specific circulating biomarkers in pediatric SCD in Ghana (West Africa), hypothesizing that inflammatory and neuronal injury markers in children with SCD could predict disease outcomes.
Methods: Clinical data were collected from 377 children aged 3-8 years with various Hb genotypes, including SCD and SCT, at Korle-Bu Teaching Hospital in Accra, Ghana (2021-2022). A total of 80 age- and sex-matched subjects were identified. A cross-sectional study utilized a multiplexed immunoassay procedure to evaluate serum biomarkers, including cytokines, chemokines, vascular injury markers, systemic inflammation markers, cell-free heme scavengers, brain-derived neurotrophic factor (BDNF), and angiogenic factors.
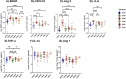
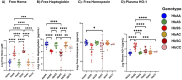
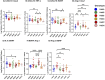
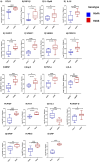
Results: Elevated levels of BDNF, Ang-2, CXCL10, CCL11, TNF-α, IL-6, IL-10, IL12p40, ICAM-1, VCAM-1, Tie-2, and VEGFA were observed in HbSS subjects, correlating with hemoglobin level, leukocyte, and erythrocyte counts. Heme scavengers like HO-1, hemopexin, and haptoglobin also correlated with these parameters. ROC and AUC analyses demonstrated the potential of these biomarkers in predicting SCD outcomes.
Conclusion: These findings suggest that there are significant differences between biomarker expression among the different genotypes examined. We conclude that a predictive algorithm based on these biomarkers could be developed and validated through longitudinal assessment of within-genotype differences and correlation of the data with disease severity or outcomes. With such a tool one can enhance SCD management and improve patient outcomes. This approach may pave the way for personalized interventions and better clinical care for pediatric SCD patients.
Keywords: global health; hemoglobinopathies; inflammation biomarkers; oxidative stress; pediatric hematology.
Copyright © 2024 Lekpor, Botchway, Driss, Bashi, Abrahams, Kusi, Futagbi, Alema-Mensah, Agbozo, Solomon, Harbuzariu, Adjei and Stiles.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Hemoglobin Genotypes Modulate Inflammatory Response to Plasmodium Infection.Front Immunol. 2020 Dec 23;11:593546. doi: 10.3389/fimmu.2020.593546. eCollection 2020. Front Immunol. 2020. PMID: 33424841 Free PMC article.
-
Assessment of Hemoglobin Variants in Patients Receiving Health Care at the Ho Teaching Hospital: A Three-Year Retrospective Study.Adv Hematol. 2020 Mar 21;2020:7369731. doi: 10.1155/2020/7369731. eCollection 2020. Adv Hematol. 2020. PMID: 32256599 Free PMC article.
-
MicroRNAs miR-451a and Let-7i-5p Profiles in Circulating Exosomes Vary among Individuals with Different Sickle Hemoglobin Genotypes and Malaria.J Clin Med. 2022 Jan 19;11(3):500. doi: 10.3390/jcm11030500. J Clin Med. 2022. PMID: 35159951 Free PMC article.
-
The Red Blood Cell-Inflammation Vicious Circle in Sickle Cell Disease.Front Immunol. 2020 Mar 13;11:454. doi: 10.3389/fimmu.2020.00454. eCollection 2020. Front Immunol. 2020. PMID: 32231672 Free PMC article. Review.
-
Current challenges in the management of patients with sickle cell disease - A report of the Italian experience.Orphanet J Rare Dis. 2019 May 30;14(1):120. doi: 10.1186/s13023-019-1099-0. Orphanet J Rare Dis. 2019. PMID: 31146777 Free PMC article. Review.
References
-
- Andrawes N. G., Ismail E. A., Roshdy M. M., Ebeid F. S. E., Eissa D. S., Ibrahim A. M. (2019). Angiopoietin-2 as a marker of retinopathy in children and adolescents with sickle cell disease: relation to subclinical atherosclerosis. J. Pediatr. Hematology/Oncology 41 (5), 361–370. 10.1097/MPH.0000000000001486 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

