MVA-based SARS-CoV-2 vaccine candidates encoding different spike protein conformations induce distinct early transcriptional responses which may impact subsequent adaptive immunity
- PMID: 39749328
- PMCID: PMC11693667
- DOI: 10.3389/fimmu.2024.1500615
MVA-based SARS-CoV-2 vaccine candidates encoding different spike protein conformations induce distinct early transcriptional responses which may impact subsequent adaptive immunity
Abstract
Introduction: Vaccine platforms such as viral vectors and mRNA can accelerate vaccine development in response to newly emerging pathogens, as demonstrated during the COVID-19 pandemic. However, the differential effects of platform and antigen insert on vaccine immunogenicity remain incompletely understood. Innate immune responses induced by viral vector vaccines are suggested to have an adjuvant effect for subsequent adaptive immunity. Integrating data on both innate and adaptive immunity, systems vaccinology approaches can improve the understanding of vaccine-induced immune mechanisms.
Methods: Two vaccine candidates against SARS-CoV-2, both based on the viral vector Modified Vaccinia virus Ankara (MVA) and encoding the native (MVA-SARS-2-S) or prefusion-stabilized spike protein (MVA-SARS-2-ST), were evaluated in phase 1 clinical trials (ClinicalTrials.gov: NCT04569383, NCT04895449). Longitudinal dynamics of innate and early adaptive immune responses induced by vaccination in SARS-CoV-2-naïve individuals were analyzed based on transcriptome and flow cytometry data, in comparison to the licensed ChAd and mRNA vaccines.
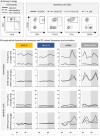
Results: Compared to MVA-SARS-2-S, MVA-SARS-2-ST (encoding the prefusion-stabilized spike protein) induced a stronger transcriptional activation early after vaccination, as well as higher virus neutralizing antibodies. Positive correlations were observed between innate and adaptive immune responses induced by a second MVA-SARS-2-ST vaccination. MVA-, ChAd- and mRNA-based vaccines induced distinct immune signatures, with the overall strongest transcriptional activation as well as monocyte and circulating T follicular helper (cTFH) cell responses induced by ChAd.
Discussion: Our findings suggest a potential impact of the spike protein conformation not only on adaptive but also on innate immune responses. As indicated by positive correlations between several immune parameters induced by MVA-SARS-2-ST, the distinct transcriptional activation early after vaccination may be linked to the induction of classical monocytes and activation of cTFH1 cells, which may in turn result in the superior adaptive immunogenicity of MVA-SARS-2-ST, compared to MVA-SARS-2-S. Overall, our data demonstrate that both the vaccine platform and antigen insert can affect innate immune responses and subsequent vaccine immunogenicity in humans.
Keywords: COVID-19; SARS-CoV-2; T follicular helper cells; innate immunity; modified vaccinia virus Ankara; spike protein; systems vaccinology; transcriptome.
Copyright © 2024 Grewe, Friedrich, Dieck, Spohn, Ly, Krähling, Mayer, Mellinghoff, Rottstegge, Kraemer, Volz, Becker, Fathi, Dahlke, Weskamm and Addo.
Conflict of interest statement
AF is an employee of BioNTech S.E. as of 01/01/2024 after completion of this study. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- EMA, E.M.A . Imvanex - smallpox and monkeypox vaccine (Live Modified Vaccinia Virus Ankara) (2022). Available online at: https://www.ema.europa.eu/en/medicines/human/EPAR/imvanex. (accessed June 30, 2024)
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

