Whole genome sequencing analysis of enteropathogenic Escherichia coli from human and companion animals in Korea
- PMID: 39749377
- PMCID: PMC11799085
- DOI: 10.4142/jvs.24225
Whole genome sequencing analysis of enteropathogenic Escherichia coli from human and companion animals in Korea
Abstract
Importance: This study is essential for comprehending the zoonotic transmission, antimicrobial resistance, and genetic diversity of enteropathogenic Escherichia coli (EPEC).
Objective: To improve our understanding of EPEC, this study focused on analyzing and comparing the genomic characteristics of EPEC isolates from humans and companion animals in Korea.
Methods: The whole genome of 26 EPEC isolates from patients with diarrhea and 20 EPEC isolates from companion animals in Korea were sequenced using the Illumina HiSeq X (Illumina, USA) and Oxford Nanopore MinION (Oxford Nanopore Technologies, UK) platforms.
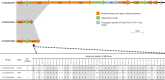
Results: Most isolates were atypical EPEC, and did not harbor the bfpA gene. The most prevalent virulence genes were found to be ompT (humans: 61.5%; companion animals: 60.0%) followed by lpfA (humans: 46.2%; companion animals: 60.0%). Although pan-genome analyses showed no apparent correlation among the origin of the strains, virulence profiles, and antimicrobial resistance profiles, isolates included in clade A obtained from both humans and companion animals exhibited high similarity. Additionally, all the isolates included in clade A encoded the ompT gene and did not encode the hlyE gene. The two isolates from companion animals harbored an incomplete bundle-forming pilus region encoding bfpA and bfpB. Moreover, the type IV secretion system-associated genes tra and trb were found in the bfpA-encoding isolates from humans.
Conclusions and relevance: Whole-genome sequencing enabled a more accurate analysis of the phylogenetic structure of EPEC and provided better insights into the understanding of EPEC epidemiology and pathogenicity.
Keywords: Enteropathogenic Escherichia coli; companion animal; human; one health; whole genome sequencing.
© 2025 The Korean Society of Veterinary Science.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Leimbach A, Hacker J, Dobrindt U. Between pathogenicity and commensalism. Curr Top Microbiol Immunol. 2013;358:3–32. - PubMed
-
- Croxen MA, Finlay BB. Molecular mechanisms of Escherichia coli pathogenicity. Nat Rev Microbiol. 2010;8(1):26–38. - PubMed
-
- Vila J, Sáez-López E, Johnson JR, Römling U, Dobrindt U, Cantón R, et al. Escherichia coli: an old friend with new tidings. FEMS Microbiol Rev. 2016;40(4):437–463. - PubMed
-
- Ledwaba SE, Bolick DT, de Medeiros PH, Kolling GL, Traore AN, Potgieter N, et al. Enteropathogenic Escherichia coli (EPEC) expressing a non-functional bundle-forming pili (BFP) also leads to increased growth failure and intestinal inflammation in C57BL/6 mice. Braz J Microbiol. 2022;53(4):1781–1787. - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

