Novel digital droplet inverse PCR assay shows that natural clearance of hepatitis B infection is associated with fewer viral integrations
- PMID: 39749570
- PMCID: PMC11731057
- DOI: 10.1080/22221751.2025.2450025
Novel digital droplet inverse PCR assay shows that natural clearance of hepatitis B infection is associated with fewer viral integrations
Abstract
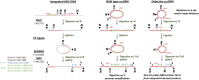
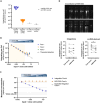
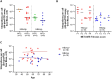
Hepatitis B virus (HBV) DNA integration into the host cell genome is reportedly a major cause of liver cancer, and a source of hepatitis B surface antigen (HBsAg). High HBsAg levels can alter immune responses which therefore contributes to the progression of HBV-related disease. However, to what extent integration leads to the persistent circulating HBsAg is unclear. Here, we aimed to determine if the extent of HBV DNA integration is associated with the persistence of circulating HBsAg in people exposed to HBV. We established a digital droplet quantitative inverse PCR (dd-qinvPCR) method to quantify integrated HBV DNA in patients who had been exposed to HBV (anti-HBc positive and HBeAg-negative). Total DNA extracts from both liver resections (n = 32; 14 HBsAg-negative and 18 HBsAg-positive) and fine-needle aspirates (FNA, n = 10; 2 HBsAg-negative and 8 HBsAg-positive) were analysed. Using defined in vitro samples for assay establishment, we showed that dd-qinvPCR could detect integrations within an input of <80 cells. The frequency of integrated HBV DNA in those who had undergone HBsAg loss (n = 14, mean ± SD of 1.514 × 10-3 ± 1.839 × 10-3 integrations per cell) was on average 9-fold lower than those with active HBV infection (n = 18, 1.16 × 10-2 ± 1.76 × 10-2 integrations per cell; p = 0.0179). In conclusion, we have developed and validated a highly precise, sensitive and quantitative PCR-based method for the quantification of HBV integrations in clinical samples. Natural clearance of HBV is associated with fewer viral integrations. Future studies are needed to determine if dynamics of integrated HBV DNA can inform the development of curative therapies.
Keywords: HBV DNA integration; HBsAg; digital droplet PCR; fine needle aspiration; functional cure.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures

References
-
- Global hepatitis report 2024: action for access in low- and middle-income countries. States News Service. 2024.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
