LGR4 promotes proliferation and homing via activation of the NF‑κB signaling pathway in multiple myeloma
- PMID: 39749708
- PMCID: PMC11753772
- DOI: 10.3892/ijo.2025.5718
LGR4 promotes proliferation and homing via activation of the NF‑κB signaling pathway in multiple myeloma
Abstract
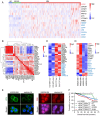
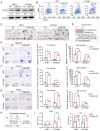
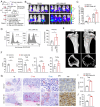
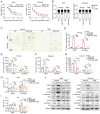
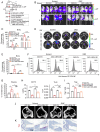
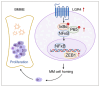
Multiple myeloma (MM) is a plasma cell malignancy characterized by clonal proliferation in the bone marrow (BM). Previously, it was reported that G‑protein‑coupled receptor 4 (LGR4) contributed to early hematopoiesis and was associated with poor prognosis in patients with MM. However, the mechanism of cell homing and migration, which is critical for MM progression, remains unclear. In the present study, cell counting, cell cycle and BrdU assays were performed to evaluate cell proliferation. Transwell assay and Xenograft mouse models were performed to evaluate cell migration and homing ability both in vitro and in vivo. I was found that overexpression of LGR4 promotes MM cell adhesion, migration and homing to BM both in vitro, while exacerbating osteolytic bone destruction in vivo. However, the LGR4 knockdown displayed the opposite effect. Further mechanistic studies demonstrated that LGR4 activated the nuclear factor kappa B (NF‑κB) signaling pathway and migration‑related adhesion molecule, thus promoting MM cell homing. Moreover, inhibiting the NF‑κB pathway was found to suppress MM cell homing. These findings identify LGR4 as a critical regulator of myeloma cell migration, homing and tumorigenesis, offering a potential therapeutic strategy for MM treatment.
Keywords: LGR4; cell homing; multiple myeloma.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Chauhan D, Uchiyama H, Akbarali Y, Urashima M, Yamamoto K, Libermann TA, Anderson KC. Multiple myeloma cell adhesion-induced interleukin-6 expression in bone marrow stromal cells involves activation of NF-kappa B. Blood. 1996;87:1104–1112. doi: 10.1182/blood.V87.3.1104.bloodjournal8731104. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
