A long noncoding RNA with enhancer-like function in pig zygotic genome activation
- PMID: 39749834
- PMCID: PMC12210014
- DOI: 10.1093/jmcb/mjae061
A long noncoding RNA with enhancer-like function in pig zygotic genome activation
Abstract
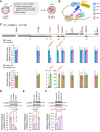
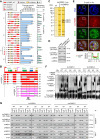
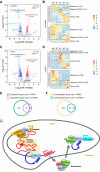
The zygotic genome activation (ZGA) is crucial for the development of pre-implantation embryos. Long noncoding RNAs (lncRNAs) play significant roles in many biological processes, but the study on their role in the early embryonic development of pigs is limited. In this study, we identify lncFKBPL as an enhancer-type lncRNA essential for pig embryo development. lncFKBPL is expressed from the 4-cell stage to the morula stage in pig embryos, and interference with lncFKBPL leads to a developmental arrest at the 8-cell stage. Mechanistic investigations uncover that lncFKBPL is able to bind to MED8, thereby mediating enhancer activity and regulating FKBPL expression. Additionally, FKBPL interacts with the molecular chaperone protein HSP90AA1, stabilizing CDK9 and boosting its protein-level expression. Elevated CDK9 levels enhance Pol II phosphorylation, facilitating ZGA. Our findings illuminate the role of lncFKBPL as an enhancer lncRNA in pig ZGA regulation and early embryo development, providing a foundation for further exploration in this area.
Keywords: lncRNA; porcine; pre-implantation embryonic development; zygotic genome activation.
© The Author(s) (2025). Published by Oxford University Press on behalf of Journal of Molecular Cell Biology, CEMCS, CAS.
Figures



Similar articles
-
Super RNA Pol II domains enhance minor ZGA through 3D interaction to ensure the integrity of major transcriptional waves in late-ZGA mammals.Cell Genom. 2025 Jun 11;5(6):100856. doi: 10.1016/j.xgen.2025.100856. Epub 2025 May 1. Cell Genom. 2025. PMID: 40315839 Free PMC article.
-
Chip (Ldb1) is a putative cofactor of Zelda forming a functional bridge to CBP during zygotic genome activation.Mol Cell. 2025 Jun 19;85(12):2425-2441.e9. doi: 10.1016/j.molcel.2025.05.018. Epub 2025 Jun 9. Mol Cell. 2025. PMID: 40494353
-
IRE1α RNase activity is critical for early embryo development by degrading maternal transcripts.Nucleic Acids Res. 2025 Jun 6;53(11):gkaf520. doi: 10.1093/nar/gkaf520. Nucleic Acids Res. 2025. PMID: 40530690 Free PMC article.
-
A meta-analysis of genome-wide association studies to identify candidate genes associated with feed efficiency traits in pigs.J Anim Sci. 2025 Jan 4;103:skaf010. doi: 10.1093/jas/skaf010. J Anim Sci. 2025. PMID: 39847436 Free PMC article.
-
Moving toward totipotency: the molecular and cellular features of totipotent and naive pluripotent stem cells.Hum Reprod Update. 2025 Jul 1;31(4):361-391. doi: 10.1093/humupd/dmaf006. Hum Reprod Update. 2025. PMID: 40299455 Review.
Cited by
-
RNA binding protein HuR regulated by OIP5-AS1 may be involved in maternal transcript degradation during the human maternal-to-zygotic transition.BMC Genomics. 2025 Jul 1;26(1):600. doi: 10.1186/s12864-025-11807-3. BMC Genomics. 2025. PMID: 40597633 Free PMC article.
References
-
- Bou G., Guo J., Liu S. et al. (2022). OCT4 expression transactivated by GATA protein is essential for non-rodent trophectoderm early development. Cell Rep. 41, 111644. - PubMed
-
- Bou G., Liu S., Sun M. et al. (2017). CDX2 is essential for cell proliferation and polarity in porcine blastocysts. Development 144, 1296–1306. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

