A Single Bioorthogonal Reaction for Multiplex Cell Surface Protein Labeling
- PMID: 39749835
- PMCID: PMC11744750
- DOI: 10.1021/jacs.4c11701
A Single Bioorthogonal Reaction for Multiplex Cell Surface Protein Labeling
Abstract
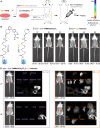
Small-molecule fluorophores are invaluable tools for fluorescence imaging. However, means for their covalent conjugation to the target proteins limit applications in multicolor imaging. Here, we identify 2-[(alkylthio)(aryl)methylene]malononitrile (TAMM) molecules reacting with 1,2-aminothiol at a labeling rate over 104 M-1 s-1 through detailed mechanistic investigation. The fast TAMM molecules and mild reaction conditions enable site-specific labeling of surface proteins in not only cell lines but also primary neurons and living mice. The combination of genetic code expansion and sequence-specific proteolytic cleavage enables selective modification of three different cell surface proteins through iterative TAMM condensation. TAMM condensation is also compatible with Cu-catalyzed azide-alkyne cycloaddition and tetrazine ligation for four-color fluorescent labeling, reaching the maximum available colors of conventional confocal microscopes. Thus, bioconjugation chemistry is no longer the limiting factor for multiplex cell surface protein imaging.
Conflict of interest statement
The authors declare the following competing financial interest(s): Yang Huang, Chengyang Wu, Anjing Lu, Jingzhe Wang, Han Sun, Chuanliu Wu, Yi-Lin Wu and Yu-Hsuan Tsai are inventors of a Chinese Patent (CN117736135B) related to this work.
Figures

 ) at 25 °C and pH 7.4 correspond to
reaction with 2x. The stability of TAMM is indicated
by the half-life (t1/2) in PBS (10 mM,
pH 7.4) at 37 °C. n.d. = not determined. (C) 1,2-Aminothiol molecules
employed in mechanistic and kinetic studies. (D) A representative
TAMM condensation between 1a and model peptide 2x to form dihydrothiazole 3ax. Left: HPLC chromatograms
of the reaction mixture analyzed at the indicated time point (λmonitor = 280 nm). Right: time evolution of [1a] and [2x] (empty circles: data points, solid lines:
fitting curves to the model of second-order reaction). (E) Effect
of TAMM para substitution (R1) on reaction
kinetics. Hammett plot of rate constants for the consumption of 1 and 2x versus the Hammett substituent constants
σp at 25 °C; slope ρ = 0.93. (F) Formation
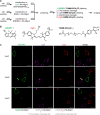
of an intermediate in TAMM condensation. HPLC chromatograms for reactions
between 1a–1d and 2x at 42 min. Peak of the intermediates is indicated with a black circle
with the m/z value of the quasi
molecular ion (either as [M + H]+ or [M–H]−) shown. (G) Formation of dihydrothiazole 3ay from disulfide 9ay by TCEP reduction. Left: HPLC chromatograms for the reaction
mixture analyzed at the time specified. Right: time course of the
concentrations of 7ay and 3ay (empty circles:
data points, solid lines: fitting curves to the model of first-order
kinetics). (H) Reaction of TAMM 1i with peptide 2x to form dihydrothiazole 3ax. Left: HPLC chromatograms
at the time specified. Right: time course of the HPLC peak intensities.
The areas under the peaks, as opposed to the concentrations, are shown
here as the calibration of Int-1, an unstable intermediate,
is not available. The dashed lines connecting data points serve as
a guide to the eye. (I) Formation of Int-2 (10) for fast-reacting TAMM supported by mass spectrum of 10ax for reactions involving 1i with peptide 2x.
) at 25 °C and pH 7.4 correspond to
reaction with 2x. The stability of TAMM is indicated
by the half-life (t1/2) in PBS (10 mM,
pH 7.4) at 37 °C. n.d. = not determined. (C) 1,2-Aminothiol molecules
employed in mechanistic and kinetic studies. (D) A representative
TAMM condensation between 1a and model peptide 2x to form dihydrothiazole 3ax. Left: HPLC chromatograms
of the reaction mixture analyzed at the indicated time point (λmonitor = 280 nm). Right: time evolution of [1a] and [2x] (empty circles: data points, solid lines:
fitting curves to the model of second-order reaction). (E) Effect
of TAMM para substitution (R1) on reaction
kinetics. Hammett plot of rate constants for the consumption of 1 and 2x versus the Hammett substituent constants
σp at 25 °C; slope ρ = 0.93. (F) Formation
of an intermediate in TAMM condensation. HPLC chromatograms for reactions
between 1a–1d and 2x at 42 min. Peak of the intermediates is indicated with a black circle
with the m/z value of the quasi
molecular ion (either as [M + H]+ or [M–H]−) shown. (G) Formation of dihydrothiazole 3ay from disulfide 9ay by TCEP reduction. Left: HPLC chromatograms for the reaction
mixture analyzed at the time specified. Right: time course of the
concentrations of 7ay and 3ay (empty circles:
data points, solid lines: fitting curves to the model of first-order
kinetics). (H) Reaction of TAMM 1i with peptide 2x to form dihydrothiazole 3ax. Left: HPLC chromatograms
at the time specified. Right: time course of the HPLC peak intensities.
The areas under the peaks, as opposed to the concentrations, are shown
here as the calibration of Int-1, an unstable intermediate,
is not available. The dashed lines connecting data points serve as
a guide to the eye. (I) Formation of Int-2 (10) for fast-reacting TAMM supported by mass spectrum of 10ax for reactions involving 1i with peptide 2x.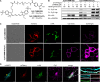

References
-
- Montecinos-Franjola F.; Bauer B. L.; Mears J. A.; Ramachandran R. GFP fluorescence tagging alters dynamin-related protein 1 oligomerization dynamics and creates disassembly-refractory puncta to mediate mitochondrial fission. Sci. Rep. 2020, 10, 14777. 10.1038/s41598-020-71655-x. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

