De novo transcriptome assembly and discovery of drought-responsive genes in white spruce (Picea glauca)
- PMID: 39752431
- PMCID: PMC11698436
- DOI: 10.1371/journal.pone.0316661
De novo transcriptome assembly and discovery of drought-responsive genes in white spruce (Picea glauca)
Abstract
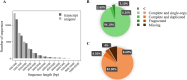
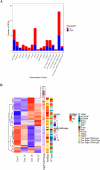
Forests face an escalating threat from the increasing frequency of extreme drought events driven by climate change. To address this challenge, it is crucial to understand how widely distributed species of economic or ecological importance may respond to drought stress. In this study, we examined the transcriptome of white spruce (Picea glauca (Moench) Voss) to identify key genes and metabolic pathways involved in the species' response to water stress. We assembled a de novo transcriptome, performed differential gene expression analyses at four time points over 22 days during a controlled drought stress experiment involving 2-year-old plants and three genetically distinct clones, and conducted gene enrichment analyses. The transcriptome assembly and gene expression analysis identified a total of 33,287 transcripts corresponding to 18,934 annotated unique genes, including 4,425 genes that are uniquely responsive to drought. Many transcripts that had predicted functions associated with photosynthesis, cell wall organization, and water transport were down-regulated under drought conditions, while transcripts linked to abscisic acid response and defense response were up-regulated. Our study highlights a previously uncharacterized effect of drought stress on lipid metabolism genes in conifers and significant changes in the expression of several transcription factors, suggesting a regulatory response potentially linked to drought response or acclimation. Our research represents a fundamental step in unraveling the molecular mechanisms underlying short-term drought responses in white spruce seedlings. In addition, it provides a valuable source of new genetic data that could contribute to genetic selection strategies aimed at enhancing the drought resistance and resilience of white spruce to changing climates.
Copyright: © 2025 Ribeyre et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Hartmann H, Bastos A, Das AJ, Esquivel-Muelbert A, Hammond WM, Martínez-Vilalta J, et al. Climate change risks to global forest health: emergence of unexpected events of elevated tree mortality worldwide. Annu Rev Plant Biol. 2022;73: 673–702. doi: 10.1146/annurev-arplant-102820-012804 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

