Antiviral activity of an ACE2-Fc fusion protein against SARS-CoV-2 and its variants
- PMID: 39752453
- PMCID: PMC11698409
- DOI: 10.1371/journal.pone.0312402
Antiviral activity of an ACE2-Fc fusion protein against SARS-CoV-2 and its variants
Abstract
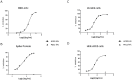
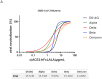
SARS-CoV-2 has continued spreading around the world in recent years since the initial outbreak in 2019, frequently developing into new variants with greater human infectious capacity. SARS-CoV-2 and its mutants use the angiotensin-converting enzyme 2 (ACE2) as a cellular entry receptor, which has triggered several therapeutic strategies against COVID-19 relying on the use of ACE2 recombinant proteins as decoy receptors. In this work, we propose an ACE2 silent Fc fusion protein (ACE2-hFcLALA) as a candidate therapy against COVID-19. This fusion protein was able to block the binding of SARS-CoV-2 RBD to ACE2 receptor as measured by ELISA and flow cytometry inhibition assays. Moreover, we used classical neutralization assays and a progeny neutralization assay to show that the ACE2-hFcLALA fusion protein is capable of neutralizing the authentic virus. Additionally, we found that this fusion protein was more effective in preventing in vitro infection with different variants of interest (alpha, beta, delta, and omicron) compared to the D614G strain. Our results suggest the potential of this molecule to be used in both therapeutic and preventive settings against current and emerging mutants that use ACE2 as a gateway to human cells.
Copyright: © 2025 Bermúdez-Abreut et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
Characterization of Entry Pathways, Species-Specific Angiotensin-Converting Enzyme 2 Residues Determining Entry, and Antibody Neutralization Evasion of Omicron BA.1, BA.1.1, BA.2, and BA.3 Variants.J Virol. 2022 Sep 14;96(17):e0114022. doi: 10.1128/jvi.01140-22. Epub 2022 Aug 24. J Virol. 2022. PMID: 36000843 Free PMC article.
-
An ACE2 Microbody Containing a Single Immunoglobulin Fc Domain Is a Potent Inhibitor of SARS-CoV-2.Cell Rep. 2020 Dec 22;33(12):108528. doi: 10.1016/j.celrep.2020.108528. Epub 2020 Dec 1. Cell Rep. 2020. PMID: 33326798 Free PMC article.
-
Humanized COVID-19 decoy antibody effectively blocks viral entry and prevents SARS-CoV-2 infection.EMBO Mol Med. 2021 Jan 11;13(1):e12828. doi: 10.15252/emmm.202012828. Epub 2020 Nov 30. EMBO Mol Med. 2021. PMID: 33159417 Free PMC article.
-
Inhibition of S-protein RBD and hACE2 Interaction for Control of SARSCoV- 2 Infection (COVID-19).Mini Rev Med Chem. 2021;21(6):689-703. doi: 10.2174/1389557520666201117111259. Mini Rev Med Chem. 2021. PMID: 33208074 Review.
-
The Repurposed ACE2 Inhibitors: SARS-CoV-2 Entry Blockers of Covid-19.Top Curr Chem (Cham). 2021 Oct 8;379(6):40. doi: 10.1007/s41061-021-00353-7. Top Curr Chem (Cham). 2021. PMID: 34623536 Free PMC article. Review.
Cited by
-
Identification and mechanism analysis of biomarkers related to butyrate metabolism in COVID-19 patients.Ann Med. 2025 Dec;57(1):2477301. doi: 10.1080/07853890.2025.2477301. Epub 2025 Mar 12. Ann Med. 2025. PMID: 40074706 Free PMC article.
References
-
- Lancet T. The COVID-19 pandemic in 2023: far from over. 2023. p. 79. - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

