Uncovering NK cell sabotage in gut diseases via single cell transcriptomics
- PMID: 39752457
- PMCID: PMC11698320
- DOI: 10.1371/journal.pone.0315981
Uncovering NK cell sabotage in gut diseases via single cell transcriptomics
Abstract
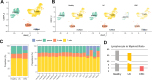
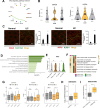
The identification of immune environments and cellular interactions in the colon microenvironment is essential for understanding the mechanisms of chronic inflammatory disease. Despite occurring in the same organ, there is a significant gap in understanding the pathophysiology of ulcerative colitis (UC) and colorectal cancer (CRC). Our study aims to address the distinct immunopathological response of UC and CRC. Using single-cell RNA sequencing datasets, we analyzed the profiles of immune cells in colorectal tissues obtained from healthy donors, UC patients, and CRC patients. The colon tissues from patients and healthy participants were visualized by immunostaining followed by laser confocal microscopy for select targets. Natural killer (NK) cells from UC patients on medication showed reduced cytotoxicity compared to those from healthy individuals. Nonetheless, a UC-specific pathway called the BAG6-NCR3 axis led to higher levels of inflammatory cytokines and increased the cytotoxicity of NCR3+ NK cells, thereby contributing to the persistence of colitis. In the context of colorectal cancer (CRC), both NK cells and CD8+ T cells exhibited significant changes in cytotoxicity and exhaustion. The GALECTIN-9 (LGALS9)-HAVCR2 axis was identified as one of the CRC-specific pathways. Within this pathway, NK cells solely communicated with myeloid cells under CRC conditions. HAVCR2+ NK cells from CRC patients suppressed NK cell-mediated cytotoxicity, indicating a reduction in immune surveillance. Overall, we elucidated the comprehensive UC and CRC immune microenvironments and NK cell-mediated immune responses. Our findings can aid in selecting therapeutic targets that increase the efficacy of immunotherapy.
Copyright: © 2025 Lee et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

