Enhanced glycolysis-derived lactate promotes microglial activation in Parkinson's disease via histone lactylation
- PMID: 39753581
- PMCID: PMC11698869
- DOI: 10.1038/s41531-024-00858-0
Enhanced glycolysis-derived lactate promotes microglial activation in Parkinson's disease via histone lactylation
Abstract
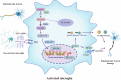
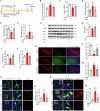
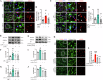
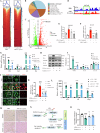
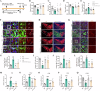
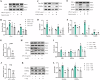
The switch from oxidative phosphorylation to glycolysis is crucial for microglial activation. Recent studies highlight that histone lactylation promotes macrophage homeostatic gene expression via transcriptional regulation, but its role in microglia activation in Parkinson's disease (PD) remains unclear. Here, we demonstrated that inhibiting glycolysis with 2-deoxy-D-glucose alleviates microgliosis, neuroinflammation and dopaminergic neurons damage by reducing lactate accumulation in PD mice. Notably, we observed a marked increase in histone lactylation, particularly H3K9 lactylation, in microglia in the substantia nigra of PD mice. Mechanistically, CUT&Tag and Chip-qPCR analyses revealed that H3K9 lactylation enriched at the SLC7A11promoter and activated its expression. Importantly, inhibiting SLC7A11 by sulfasalazine mitigated microglia-mediated neuroinflammation and improved motor function in PD mice. Moreover, we found that lactate-induce histone lactylation is dependent on P300/CBP. Collectively, our findings demonstrate that glycolysis-derived lactate promotes microglial activation via histone lactylation and provide a potential therapeutic strategy for PD.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
-
- Kalia, L. V. & Lang, A. E. Parkinson’s disease [J]. Lancet386, 896–912 (2015). - PubMed
-
- Stokholm, M. G. et al. Assessment of neuroinflammation in patients with idiopathic rapid-eye-movement sleep behaviour disorder: a case-control study [J]. Lancet Neurol.16, 789–796 (2017). - PubMed
-
- Long-Smith, C. M., Sullivan, A. M. & Nolan, Y. M. The influence of microglia on the pathogenesis of Parkinson’s disease [J]. Prog. Neurobiol.89, 277–287 (2009). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

