Bacteria use exogenous peptidoglycan as a danger signal to trigger biofilm formation
- PMID: 39753671
- PMCID: PMC11726461
- DOI: 10.1038/s41564-024-01886-5
Bacteria use exogenous peptidoglycan as a danger signal to trigger biofilm formation
Abstract
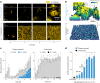
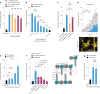
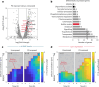
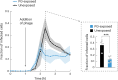
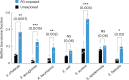
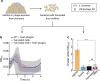
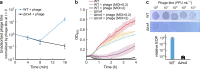
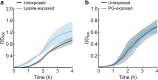
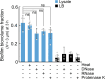
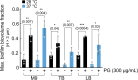
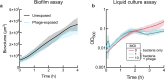
For any organism, survival is enhanced by the ability to sense and respond to threats in advance. For bacteria, danger sensing among kin cells has been observed, but the presence or impacts of general danger signals are poorly understood. Here we show that different bacterial species use exogenous peptidoglycan fragments, which are released by nearby kin or non-kin cell lysis, as a general danger signal. Using microscopy and gene expression profiling of Vibrio cholerae, we find that even brief signal exposure results in a regulatory response that causes three-dimensional biofilm formation, which protects cells from a broad range of stresses, including bacteriophage predation. A diverse set of species (Pseudomonas aeruginosa, Acinetobacter baumannii, Staphylococcus aureus, Enterococcus faecalis) also respond to exogenous peptidoglycan by forming biofilms. As peptidoglycan from different Gram-negative and Gram-positive species triggered three-dimensional biofilm formation, we propose that this danger signal and danger response are conserved among bacteria.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures





References
MeSH terms
Substances
Grants and funding
- TMCG-3_213801/Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung (Swiss National Science Foundation)
- 51NF40_180541/Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung (Swiss National Science Foundation)
- DR 982/5-1/Deutsche Forschungsgemeinschaft (German Research Foundation)
- DR 982/6-1/Deutsche Forschungsgemeinschaft (German Research Foundation)
- TARGET-Biofilms/Bundesministerium für Bildung und Forschung (Federal Ministry of Education and Research)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

