Comprehensive genomic characterization of early-stage bladder cancer
- PMID: 39753772
- PMCID: PMC11735393
- DOI: 10.1038/s41588-024-02030-z
Comprehensive genomic characterization of early-stage bladder cancer
Abstract
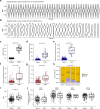
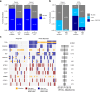
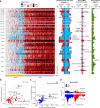
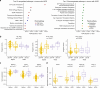
Understanding the molecular landscape of nonmuscle-invasive bladder cancer (NMIBC) is essential to improve risk assessment and treatment regimens. We performed a comprehensive genomic analysis of patients with NMIBC using whole-exome sequencing (n = 438), shallow whole-genome sequencing (n = 362) and total RNA sequencing (n = 414). A large genomic variation within NMIBC was observed and correlated with different molecular subtypes. Frequent loss of heterozygosity in FGFR3 and 17p (affecting TP53) was found in tumors with mutations in FGFR3 and TP53, respectively. Whole-genome doubling (WGD) was observed in 15% of the tumors and was associated with worse outcomes. Tumors with WGD were genomically unstable, with alterations in cell-cycle-related genes and an altered immune composition. Finally, integrative clustering of multi-omics data highlighted the important role of genomic instability and immune cell exhaustion in disease aggressiveness. These findings advance our understanding of genomic differences associated with disease aggressiveness in NMIBC and may ultimately improve patient stratification.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures










References
-
- Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin.71, 209–249 (2021). - PubMed
-
- Babjuk, M. et al. European Association of Urology Guidelines on non-muscle-invasive bladder cancer (Ta, T1, and carcinoma in situ). Eur. Urol.81, 75–94 (2022). - PubMed
-
- Dyrskjøt, L. & Ingersoll, M. A. Biology of nonmuscle-invasive bladder cancer: pathology, genomic implications, and immunology. Curr. Opin. Urol.28, 598–603 (2018). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

