ATM and IRAK1 orchestrate two distinct mechanisms of NF-κB activation in response to DNA damage
- PMID: 39753776
- PMCID: PMC11997730
- DOI: 10.1038/s41594-024-01417-0
ATM and IRAK1 orchestrate two distinct mechanisms of NF-κB activation in response to DNA damage
Erratum in
-
Publisher Correction: ATM and IRAK1 orchestrate two distinct mechanisms of NF-κB activation in response to DNA damage.Nat Struct Mol Biol. 2025 Aug;32(8):1580. doi: 10.1038/s41594-025-01638-x. Nat Struct Mol Biol. 2025. PMID: 40624165 No abstract available.
Abstract
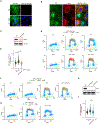
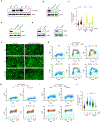
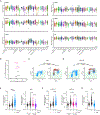
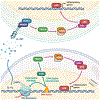
DNA damage in cells induces the expression of inflammatory genes. However, the mechanism by which cells initiate an innate immune response in the presence of DNA lesions blocking transcription remains unknown. Here we find that genotoxic stresses lead to an acute activation of the transcription factor NF-κB through two distinct pathways, each triggered by different types of DNA lesions and coordinated by either ataxia-telangiectasia mutated (ATM) or IRAK1 kinases. ATM stimulates NF-κB in cells with DNA double-strand breaks. By contrast, IRAK1-induced NF-κB signaling occurs in neighboring cells through IL-1α secretion from transcriptionally stressed cells caused by DNA lesions blocking RNA polymerases. Subsequently, both pathways stimulate TRAF6 and the IKK complex to promote NF-κB-mediated inflammatory gene expression. These findings provide an alternative mechanism for damaged cells with impaired transcription to initiate an inflammatory response without relying on their own gene expression, a necessary step that injured cells depend on during canonical innate immune responses.
© 2025. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures
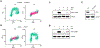


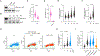






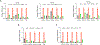







References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

