Chaperone-mediated insertion of mitochondrial import receptor TOM70 protects against diet-induced obesity
- PMID: 39753947
- PMCID: PMC12117470
- DOI: 10.1038/s41556-024-01555-z
Chaperone-mediated insertion of mitochondrial import receptor TOM70 protects against diet-induced obesity
Abstract
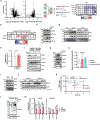
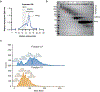
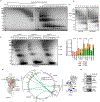
Outer mitochondrial membrane (OMM) proteins communicate with the cytosol and other organelles, including the endoplasmic reticulum. This communication is important in thermogenic adipocytes to increase the energy expenditure that controls body temperature and weight. However, the regulatory mechanisms of OMM protein insertion are poorly understood. Here the stress-induced cytosolic chaperone PPID (peptidyl-prolyl isomerase D/cyclophilin 40/Cyp40) drives OMM insertion of the mitochondrial import receptor TOM70 that regulates body temperature and weight in obese mice, and respiratory/thermogenic function in brown adipocytes. PPID PPIase activity and C-terminal tetratricopeptide repeats, which show specificity towards TOM70 core and C-tail domains, facilitate OMM insertion. Our results provide an unprecedented role for endoplasmic-reticulum-stress-activated chaperones in controlling energy metabolism through a selective OMM protein insertion mechanism with implications in adaptation to cold temperatures and high-calorie diets.
© 2025. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures










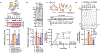


References
Methods-only references
MeSH terms
Substances
Grants and funding
- F32 GM125243/GM/NIGMS NIH HHS/United States
- R01 DK097441/DK/NIDDK NIH HHS/United States
- 24POST1198357/American Heart Association (American Heart Association, Inc.)
- 1T32GM145407/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- R01 DK081418/DK/NIDDK NIH HHS/United States
- R35 CA242461/CA/NCI NIH HHS/United States
- R01DK136640/U.S. Department of Health & Human Services | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (National Institute of Diabetes & Digestive & Kidney Diseases)
- R01 DK125283/DK/NIDDK NIH HHS/United States
- R01 DK136640/DK/NIDDK NIH HHS/United States
- GM125243/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- R01 DK089883/DK/NIDDK NIH HHS/United States
- P30 DK135043/DK/NIDDK NIH HHS/United States
- S10 OD028635/OD/NIH HHS/United States
- T32 GM145407/GM/NIGMS NIH HHS/United States
- K99 DK133502/DK/NIDDK NIH HHS/United States
- R01 DK138529/DK/NIDDK NIH HHS/United States
- R01 DK133948/DK/NIDDK NIH HHS/United States
- DP1 DK126160/DK/NIDDK NIH HHS/United States
- K99DK133502/U.S. Department of Health & Human Services | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (National Institute of Diabetes & Digestive & Kidney Diseases)
- R01DK081418/U.S. Department of Health & Human Services | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (National Institute of Diabetes & Digestive & Kidney Diseases)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

