Personalized, autologous neoantigen-specific T cell therapy in metastatic melanoma: a phase 1 trial
- PMID: 39753970
- PMCID: PMC11922764
- DOI: 10.1038/s41591-024-03418-4
Personalized, autologous neoantigen-specific T cell therapy in metastatic melanoma: a phase 1 trial
Erratum in
-
Publisher Correction: Personalized, autologous neoantigen-specific T cell therapy in metastatic melanoma: a phase 1 trial.Nat Med. 2025 Sep;31(9):3206. doi: 10.1038/s41591-025-03896-0. Nat Med. 2025. PMID: 40760355 Free PMC article. No abstract available.
Abstract
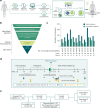
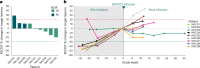
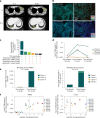
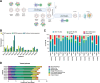
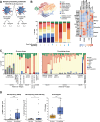
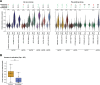
New treatment approaches are warranted for patients with advanced melanoma refractory to immune checkpoint blockade (ICB) or BRAF-targeted therapy. We designed BNT221, a personalized, neoantigen-specific autologous T cell product derived from peripheral blood, and tested this in a 3 + 3 dose-finding study with two dose levels (DLs) in patients with locally advanced or metastatic melanoma, disease progression after ICB, measurable disease (Response Evaluation Criteria in Solid Tumors version 1.1) and, where appropriate, BRAF-targeted therapy. Primary and secondary objectives were evaluation of safety, highest tolerated dose and anti-tumor activity. We report here the non-pre-specified, final results of the completed monotherapy arm consisting of nine patients: three at DL1 (1 × 108-1 × 109 cells) and six at DL2 (2 × 109-1 × 1010 cells). Drug products (DPs) were generated for all enrolled patients. BNT221 was well tolerated across both DLs, with no dose-limiting toxicities of grade 3 or higher attributed to the T cell product observed. Specifically, no cytokine release, immune effector cell-associated neurotoxicity or macrophage activation syndromes were reported. A dose of 5.0 × 108-1.0 × 1010 cells was identified for further study conduct. Six patients showed stable disease as best overall response, and tumor reductions (≤20%) were reported for four of these patients. In exploratory analyses, multiple mutant-specific CD4+ and CD8+ T cell responses were generated in each DP. These were cytotoxic, polyfunctional and expressed T cell receptors with broad functional avidities. Neoantigen-specific clonotypes were detected after treatment in blood and tumor. Our results provide key insights into this neoantigen-specific adoptive T cell therapy and demonstrate proof of concept for this new therapeutic approach. ClinicalTrials.gov registration: NCT04625205 .
© 2025. The Author(s).
Conflict of interest statement
Competing interests: J.S.W.B., J.V.v.T., S.K., S.W. and I.J. declare no competing interests. D.L., V.K., E.K.J., S.H., E.O.C., K.N.B., E.E., K.S., Y.W., T.S., X.C., G.M., J.H.F.S., S.R., M.R., J.Z.D., J.R.S., V.R.J., C.M.A., C.G., O.C.F., K.M., R.B.G., M.D.M. and M.M.v.B. are employees of BioNTech US or BioNTech SE. B.M.C. is a former employee of BioNTech US. V.K., S.H., K.N.B., K.S., Y.W., X.C., S.R., J.R.S., V.R.J., C.M.A, O.C.F., K.M., R.B.G., M.D.M. and M.M.v.B. own stock in BioNTech. C.M.N., B.N., K.C.S. and M.D.L. are contractors for BioNTech US for manufacture of the BNT221 drug product. M.M.v.B., D.L., C.M.A., J.B.H. and M.D.M. are inventors on patent applications that cover part of this article. J.B.H. received grant or research support from BioNTech US, Bristol Myers Squibb, Novartis AG, Amgen, Sastra Cell Therapy and Asher Bio. BioNTech’s role in this study is disclosed in the author contributions; none of the other organizations had a role in the study or publication. J.B.H. discloses ownership interest in Neogene Therapeutics. J.B.H. has an advisory role for Agenus, AstraZeneca, Bristol Myers Squibb, CureVac, GSK, Imcyse, Iovance Bio, Immunocore, Ipsen, Merck Serono, Merck Sharp & Dohme, Molecular Partners, Novartis, Orgenesis, Pfizer, Roche/Genentech, Sanofi and Third Rock Ventures and is a scientific advisory board (SAB) member at Achilles Tx, BioNTech US, Instil Bio, T-Knife, Neogene Therapeutics (AstraZeneca) and Sastra Cell Therapy. R.B.G. is on the Board of Directors for Alkermes and Zai Lab and is a member of the SAB for Leap Therapeutics.
Figures




References
-
- Michielin, O. et al. Cutaneous melanoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol.30, 1884–1901 (2019). - PubMed
-
- Seth, R. et al. Systemic therapy for melanoma: ASCO guideline update. J. Clin. Oncol.41, 4794–4820 (2023). - PubMed
-
- Larkin, J. et al. Five-year survival with combined nivolumab and ipilimumab in advanced melanoma. N. Engl. J. Med.381, 1535–1546 (2019). - PubMed
-
- Curti, B. D. & Faries, M. B. Recent advances in the treatment of melanoma. N. Engl. J. Med.384, 2229–2240 (2021). - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

