Cardiac conduction system regeneration prevents arrhythmias after myocardial infarction
- PMID: 39753976
- PMCID: PMC11825367
- DOI: 10.1038/s44161-024-00586-x
Cardiac conduction system regeneration prevents arrhythmias after myocardial infarction
Abstract
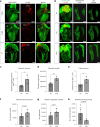
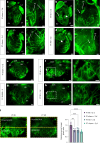
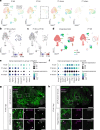
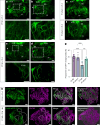
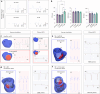
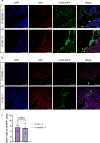
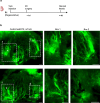
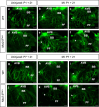
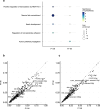
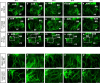
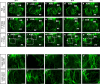
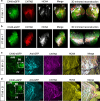
Arrhythmias are a hallmark of myocardial infarction (MI) and increase patient mortality. How insult to the cardiac conduction system causes arrhythmias following MI is poorly understood. Here, we demonstrate conduction system restoration during neonatal mouse heart regeneration versus pathological remodeling at non-regenerative stages. Tissue-cleared whole-organ imaging identified disorganized bundling of conduction fibers after MI and global His-Purkinje disruption. Single-cell RNA sequencing (scRNA-seq) revealed specific molecular changes to regenerate the conduction network versus aberrant electrical alterations during fibrotic repair. This manifested functionally as a transition from normal rhythm to pathological conduction delay beyond the regenerative window. Modeling in the infarcted human heart implicated the non-regenerative phenotype as causative for heart block, as observed in patients. These findings elucidate the mechanisms underpinning conduction system regeneration and reveal how MI-induced damage elicits clinical arrhythmogenesis.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
-
- Levine, H. J. Rest heart rate and life expectancy. J. Am. Coll. Cardiol.30, 1104–1106 (1997). - PubMed
-
- Boyden, P. A., Albala, A. & Dresdner, K. P. Jr Electrophysiology and ultrastructure of canine subendocardial Purkinje cells isolated from control and 24-hour infarcted hearts. Circ. Res.65, 955–970 (1989). - PubMed
-
- Bogun, F. et al. Role of Purkinje fibers in post-infarction ventricular tachycardia. J. Am. Coll. Cardiol.48, 2500–2507 (2006). - PubMed
MeSH terms
Grants and funding
- icp016/Partnership for Advanced Computing in Europe AISBL (PRACE)
- CAPES, CNPq, FAPEMIG/Governo Brasil (Brazilian Government)
- EP/X019446/1/RCUK | Engineering and Physical Sciences Research Council (EPSRC)
- BB/V509395/1/RCUK | Biotechnology and Biological Sciences Research Council (BBSRC)
- 214290/Z/18/Z/Wellcome Trust (Wellcome)
LinkOut - more resources
Full Text Sources
Medical
