Overcoming CD226-related immune evasion in acute myeloid leukemia with CD38 CAR-engineered NK cells
- PMID: 39754720
- PMCID: PMC11838179
- DOI: 10.1016/j.celrep.2024.115122
Overcoming CD226-related immune evasion in acute myeloid leukemia with CD38 CAR-engineered NK cells
Abstract
CD226 plays a vital role in natural killer (NK) cell cytotoxicity, interacting with its ligands CD112 and CD155 to initiate immune synapse formation, primarily through leukocyte function-associated-1 (LFA-1). Our study examined the role of CD226 in NK cell surveillance of acute myeloid leukemia (AML). NK cells in patients with AML had lower expression of CD226. CRISPR-Cas9 deletion of CD226 led to reduced LFA-1 recruitment, poor synapse formation, and decreased NK cell anti-leukemic activity. Engineering NK cells to express a chimeric antigen receptor targeting the AML antigen CD38 (CAR38) could overcome the need for CD226 to establish strong immune synapses. LFA-1 blockade reduced CAR38 NK cell activity, and this depended on the CD38 expression levels of AML cells. This suggests parallel but potentially cooperative roles for LFA-1 and CAR38 in synapse formation. Our findings suggest that CAR38 NK cells could be an effective therapeutic strategy to overcome CD226-mediated immune evasion in AML.
Keywords: CAR-NK cell; CD226; CP: Cancer; CP: Immunology; LFA-1; acute myeloid leukemia; immune synapse.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests P.B., Y.L., P.L., M.D., H.R., R.B., E.L., E.J.S., K.R., and The University of Texas MD Anderson Cancer Center have an institutional financial conflict of interest with Takeda Pharmaceutical. R.B., E.L., and K.R. have an institutional financial conflict of interest with Affimed GmbH. K.R. participates on the scientific advisory boards of Avenge Bio, Virogin Biotech, Navan Technologies, Caribou Biosciences, Bit Bio Limited, Replay Holdings, oNKo Innate, The Alliance for Cancer Gene Therapy ACGT, Innate Pharma, and Shinobi Therapeutics. K.R. is the scientific founder of Syena. E.J.S. has served on the scientific advisory boards of Adaptimmune, Axio, Celaid, FibroBiologics, Navan Technologies, New York Blood Center, and Novartis. M.D. participates on the scientific advisory board of Cellsbin.
Figures

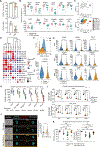


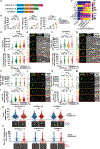

References
-
- Breems DA, Van Putten WLJ, Huijgens PC, Ossenkoppele GJ, Verhoef GEG, Verdonck LF, Vellenga E, De Greef GE, Jacky E, Van der Lelie J, et al. (2005). Prognostic index for adult patients with acute myeloid leukemia in first relapse. J. Clin. Oncol. 23, 1969–1978. 10.1200/JCO.2005.06.027. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

