Serological markers of exocrine pancreatic function are differentially informative for distinguishing individuals progressing to type 1 diabetes
- PMID: 39755561
- PMCID: PMC11749058
- DOI: 10.1136/bmjdrc-2024-004655
Serological markers of exocrine pancreatic function are differentially informative for distinguishing individuals progressing to type 1 diabetes
Abstract
Introduction: Altered serum levels of growth hormones, adipokines, and exocrine pancreas enzymes have been individually linked with type 1 diabetes (T1D). We collectively evaluated seven such biomarkers, combined with islet autoantibodies (AAb) and genetic risk score (GRS2), for their utility in predicting AAb/T1D status.
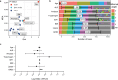
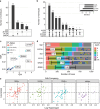
Research design and methods: Cross-sectional serum samples (n=154 T1D, n=56 1AAb+, n=77 ≥2AAb+, n=256 AAb-) were assessed for IGF1, IGF2, adiponectin, leptin, amylase, lipase, and trypsinogen (n=543, age range 2.7-30.0 years) using random forest modeling.
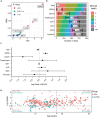
Results: GRS2, age, lipase, trypsinogen, and AAb against ZnT8, GAD65, and insulin were the most informative markers. Notably, these variables were differentially informative according to AAb/T1D status. Higher GRS2 (p<0.001) and lower lipase levels (p=0.002) favored ≥2AAb+ versus AAb- classification. AAb against ZnT8 (p<0.01), GAD65 (p=0.021), or insulin (p=0.01) each independently favored ≥2AAb+ versus 1AAb+ classification. Reduced trypsinogen (p<0.001) and increased lipase levels (p<0.001) favored recent-onset T1D versus ≥2AAb+ classification.
Conclusions: Among the serological markers tested, lipase and trypsinogen levels were the most informative for differentiating among clinical groups, with the utility of each enzyme varying according to GRS2 and AAb/T1D status. These data support exocrine pancreas enzymes as candidates for longitudinal follow-up.
Keywords: Autoantibodies; Diabetes Mellitus, Type 1; Enzymes; Models, Statistical.
© Author(s) (or their employer(s)) 2024. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ Group.
Conflict of interest statement
Competing interests: None declared.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
