Profiling of pathogenic variants in Japanese patients with sarcoglycanopathy
- PMID: 39755676
- PMCID: PMC11699685
- DOI: 10.1186/s13023-024-03521-2
Profiling of pathogenic variants in Japanese patients with sarcoglycanopathy
Abstract
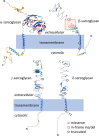
Background: Sarcoglycanopathies (SGPs) are limb-girdle muscular dystrophies (LGMDs) that can be classified into four types, LGMDR3, LGMDR4, LGMDR5, and LGMDR6, caused by mutations in the genes, SGCA, SGCB, SGCG, and SGCD, respectively. SGPs are relatively rare in Japan. This study aims to profile the genetic variants that cause SGPs in Japanese patients.
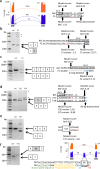
Methods: Clinical course and pathological findings were retrospectively reviewed in Japanese patients with SGP. Genetic analyses were performed using a combination of targeted resequencing with a hereditary muscle disease panel, whole genome sequencing, multiplex ligation-dependent probe amplification, and long-read sequencing. The structures of transcripts with aberrant splicing were also determined by RT-PCR, RNA-seq, and in silico prediction.
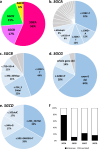
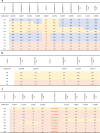
Results: We identified biallelic variants in SGC genes in 53 families, including three families with LGMDR6, which had not been identified in Japan so far. SGCA was the most common causative gene, accounting for 56% of cases, followed by SGCG, SGCB, and SGCD, at 17%, 21%, and 6%, respectively. Missense variants in SGCA were very frequent at 78.3%, while they were relatively rare in SGCB, SGCG, and SGCD at 11.1%, 18.2%, and 16.6%, respectively. We also analyzed the haplotypes of alleles carrying three variants found in multiple cases: c.229C > T in SGCA, c.325C > T in SGCB, and exon 6 deletion in SGCG; two distinct haplotypes were found for c.229C > T in SGCA, while each of the latter two variants was on single haplotypes.
Conclusions: We present genetic profiles of Japanese patients with SGPs. Haplotype analysis indicated common ancestors of frequent variants. Our findings will support genetic diagnosis and gene therapy.
Keywords: Genetic Profile; Haplotyping; Japan; Large deletion; Limb-Girdle muscular dystrophies; RNA-Seq; Sarcoglycanopathies; Skeletal muscles, in silico prediction; Variants.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: All clinical information and materials used in this study were obtained for diagnostic purposes with written informed consent. The study was approved by the Ethics Committee of the National Center of Neurology and Psychiatry (NCNP). Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures

References
-
- Straub V, Murphy A, Udd B, LGMD workshop study group. 229th ENMC international workshop: Limb girdle muscular dystrophies - Nomenclature and reformed classification Naarden. Neuromuscul Disord. 2018;28:02–10. - PubMed
-
- Alonso-Pérez J, González-Quereda L, Bello L, Guglieri M, Straub V, Gallano P, et al. New genotype-phenotype correlations in a large European cohort of patients with sarcoglycanopathy. Brain. 2020;143:2696–708. - PubMed
-
- Dalichaouche I, Sifi Y, Roudaut C, Sifi K, Hamri A, Rouabah L, et al. γ-sarcoglycan and dystrophin mutation spectrum in an Algerian cohort. Muscle Nerve. 2017;56:129–35. - PubMed
-
- Liang W-C, Chou P-C, Hung C-C, Su Y-N, Kan T-M, Chen W-Z, et al. Probable high prevalence of limb-girdle muscular dystrophy type 2D in Taiwan. J Neurol Sci. 2016;362:304–8. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

