Multi-omics data integration identifies novel biomarkers and patient subgroups in inflammatory bowel disease
- PMID: 39756419
- PMCID: PMC11792892
- DOI: 10.1093/ecco-jcc/jjae197
Multi-omics data integration identifies novel biomarkers and patient subgroups in inflammatory bowel disease
Abstract
Background: Inflammatory bowel disease (IBD), comprising Crohn's disease (CD) and ulcerative colitis (UC), is a complex condition with diverse manifestations; recent advances in multi-omics technologies are helping researchers unravel its molecular characteristics to develop targeted treatments.
Objectives: In this work, we explored one of the largest multi-omics cohorts in IBD, the Study of a Prospective Adult Research Cohort (SPARC IBD), with the goal of identifying predictive biomarkers for CD and UC and elucidating patient subtypes.
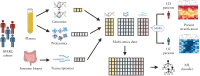
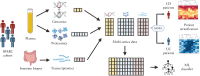
Design: We analyzed genomics, transcriptomics (gut biopsy samples), and proteomics (blood plasma) from hundreds of patients from SPARC IBD. We trained a machine learning model that classifies UC versus CD samples. In parallel, we integrated multi-omics data to unveil patient subgroups in each of the 2 indications independently and analyzed the molecular phenotypes of these patient subpopulations.
Results: The high performance of the model showed that multi-omics signatures are able to discriminate between the 2 indications. The most predictive features of the model, both known and novel omics signatures for IBD, can potentially be used as diagnostic biomarkers. Patient subgroup analysis in each indication uncovered omics features associated with disease severity in UC patients and with tissue inflammation in CD patients. This culminates with the observation of 2 CD subpopulations characterized by distinct inflammation profiles.
Conclusions: Our work unveiled potential biomarkers to discriminate between CD and UC and to stratify each population into well-defined subgroups, offering promising avenues for the application of precision medicine strategies.
Keywords: Crohn’s disease; inflammatory bowel disease; machine learning; multi-omics; precision medicine; ulcerative colitis.
© The Author(s) 2025. Published by Oxford University Press on behalf of European Crohn’s and Colitis Organisation.
Conflict of interest statement
All authors were employees of Enveda Inc. during the course of this work and have real or potential ownership interest in the company.
Figures




References
-
- Baumgart DC, Carding SR.. Inflammatory bowel disease: cause and immunobiology. Lancet. 2007;369:1627–1640. https://doi.org/ 10.1016/S0140-6736(07)60750-8 - DOI - PubMed
-
- Weersma RK, Xavier RJ, Vermeire S, Barrett JC; IBD Multi Omics Consortium. Multiomics analyses to deliver the most effective treatment to every patient with inflammatory bowel disease. Gastroenterol. 2018;155:e1–e4. https://doi.org/ 10.1053/j.gastro.2018.07.039 - DOI - PubMed
-
- Lloyd-Price J, Arze C, Ananthakrishnan AN, et al. ; IBDMDB Investigators. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569:655–662. https://doi.org/ 10.1038/s41586-019-1237-9 - DOI - PMC - PubMed
-
- Agrawal M, Allin KH, Petralia F, Colombel JF, Jess T.. Multiomics to elucidate inflammatory bowel disease risk factors and pathways. Nat Rev Gastroenterol Hepatol. 2022;19:399–409. https://doi.org/ 10.1038/s41575-022-00593-y - DOI - PMC - PubMed
-
- Subramanian I, Verma S, Kumar S, Jere A, Anamika K.. Multi-omics data integration, interpretation, and its application. Bioinf Biol Insights. 2020;14:1. https://doi.org/ 10.1177/1177932219899051 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

