A pan-immune panorama of bacterial pneumonia revealed by a large-scale single-cell transcriptome atlas
- PMID: 39757231
- PMCID: PMC11701081
- DOI: 10.1038/s41392-024-02093-8
A pan-immune panorama of bacterial pneumonia revealed by a large-scale single-cell transcriptome atlas
Abstract
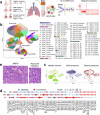
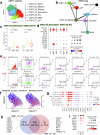
Bacterial pneumonia is a significant public health burden, contributing to substantial morbidity, mortality, and healthcare costs. Current therapeutic strategies beyond antibiotics and adjuvant therapies are limited, highlighting the need for a deeper understanding of the disease pathogenesis. Here, we employed single-cell RNA sequencing of 444,146 bronchoalveolar lavage fluid cells (BALFs) from a large cohort of 74 individuals, including 58 patients with mild (n = 22) and severe (n = 36) diseases as well as 16 healthy donors. Enzyme-linked immunosorbent and histological assays were applied for validation within this cohort. The heterogeneity of immune responses in bacterial pneumonia was observed, with distinct immune cell profiles related to disease severity. Severe bacterial pneumonia was marked by an inflammatory cytokine storm resulting from systemic upregulation of S100A8/A9 and CXCL8, primarily due to specific macrophage and neutrophil subsets. In contrast, mild bacterial pneumonia exhibits an effective humoral immune response characterized by the expansion of T follicular helper and T helper 2 cells, facilitating B cell activation and antibody production. Although both disease groups display T cell exhaustion, mild cases maintained robust cytotoxic CD8+T cell function, potentially reflecting a compensatory mechanism. Dysregulated neutrophil and macrophage responses contributed significantly to the pathogenesis of severe disease. Immature neutrophils promote excessive inflammation and suppress T cell activation, while a specific macrophage subset (Macro_03_M1) displaying features akin to myeloid-derived suppressor cells (M-MDSCs) suppress T cells and promote inflammation. Together, these findings highlight potential therapeutic targets for modulating immune responses and improving clinical outcomes in bacterial pneumonia.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
-
- Liu, Y. N. et al. Infection and co-infection patterns of community-acquired pneumonia in patients of different ages in China from 2009 to 2020: a national surveillance study. Lancet Microbe4, e330–e339 (2023). - PubMed
-
- Martin-Loeches, I. et al. ERS/ESICM/ESCMID/ALAT guidelines for the management of severe community-acquired pneumonia. Eur. Respir. J.61, 615–632 (2023). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

